|
Version: 2.1 Run ID: 20593 Run Name: mhcII_20593 Date: Tue Jan 8 11:44:16 2019 Training data• Read 700 sequencesView data distribution (See Instructions for optimal data distribution) • Pre-processing: Linear rescale |
Neural network architecture• Motif length: 9• Flanking region (PFR) size: 3 • PFR length encoding type: 0 • PFR encoding: Blosum • Expected peptide length for encoding: 15 • Number of hidden neurons: 10 • Maximum length of deletions in alignment: 0 • Maximum length of insertions in alignment: 0 • Burn-in period without insertions or deletions: 1 • Impose amino acid preference during burn-in: ILVMFYW • Amino acid numerical encoding: Blosum • Number of training cycles: 200 • Number of random seeds: 10 • Number of networks in final ensemble: 10 No cross-validation - training on all data |
RESULTS
Performance measures - motif length 9
You chose to perform no cross-validation.No performance measures are available on the training set
Save the trained MODEL. You may use this model for a new submission
Sequence motif
Cores realigned with offset correction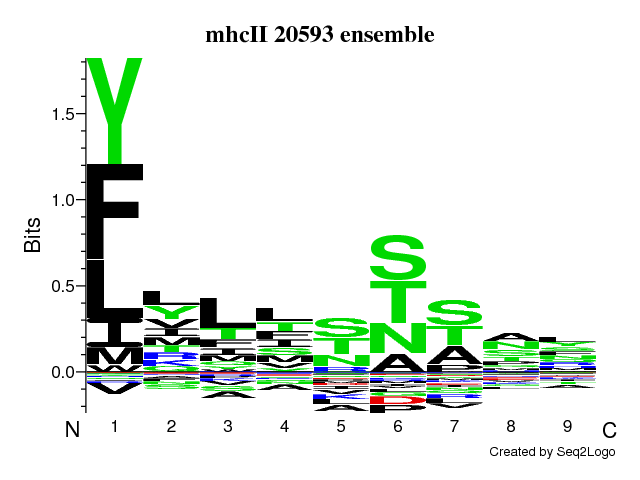
Click here if you have problems visualizing this image
Figure: Visualization of the sequence motif using the Seq2Logo program
View a Log-odds matrix or Frequency matrix representation of the motif
Evaluation data
Uploaded 800 peptidesSee the predictions on the evaluation set
RMSE = 0.20446
Pearson correlation = 0.57302
Spearman correlation = 0.58183
View scatterplot of predicted vs. observed values for evaluation set
DOWNLOAD a compressed archive with all results files
Explain the output
Go back