DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
TCRcluster is a variational autoencoder and agglomerative clustering pipeline. It clusters TCRs together based on latent cosine distances extracted with a VAE.
Click here to download the latent vector and predicted clusters in .csv format.
Click here to download the clusters summary in .csv format.
Click here to download the cosine distance matrix in .csv format.
Click here to download the optimisation results in .csv format.
Click here to download the optimisation curve plot in .png format.
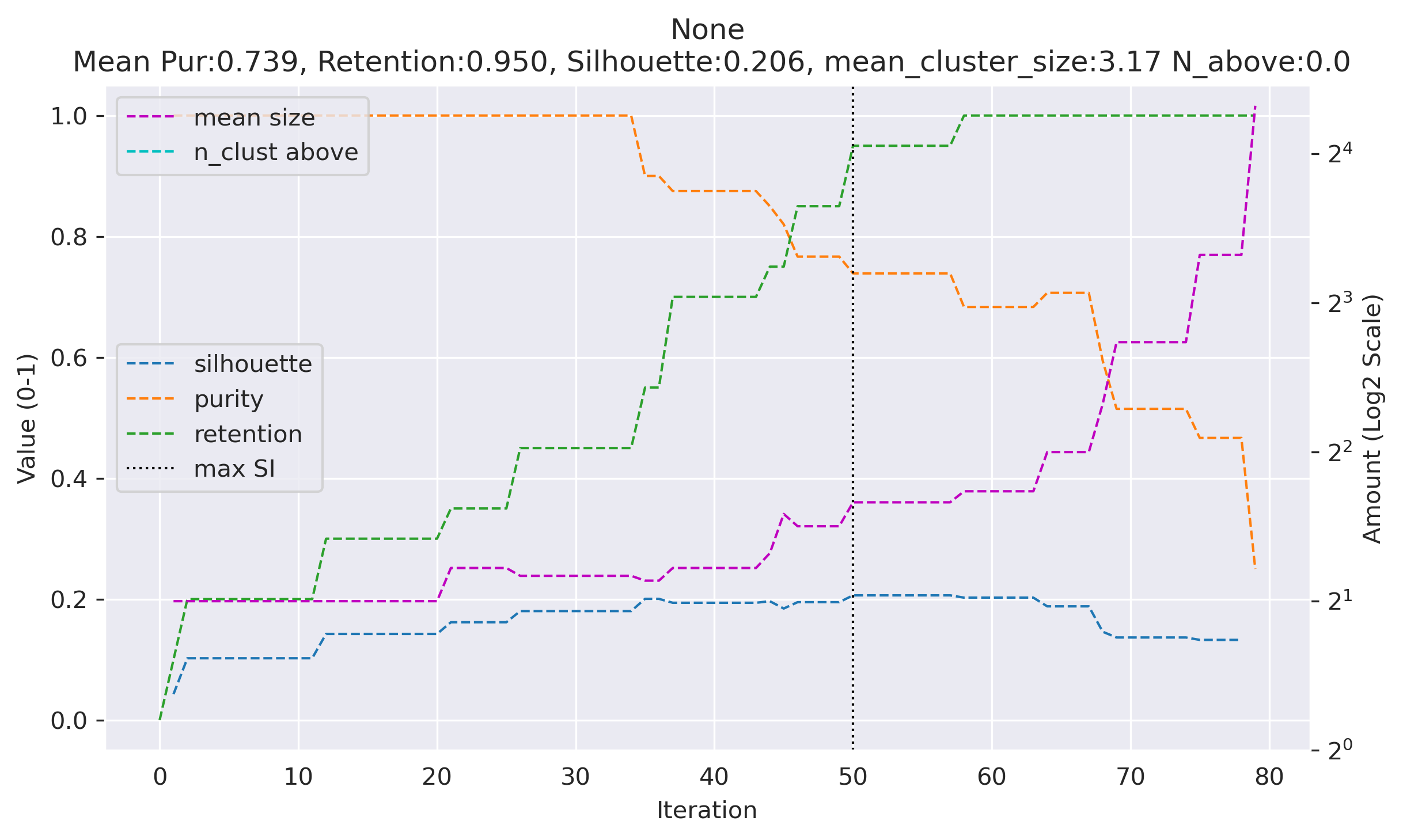
Below is a table preview of clustering metrics at each threshold tested.
A total of 80 points are tested, showing only 10 points centered around the best solution.
the 'best' column denotes the best silhouette solution.
threshold best n_cluster n_singletons silhouette mean_purity \
40 0.77949 False 6 6 0.194 0.875
41 0.79165 False 6 6 0.194 0.875
42 0.80380 False 6 6 0.194 0.875
43 0.81596 False 6 6 0.194 0.875
44 0.82811 False 6 5 0.197 0.850
45 0.84026 False 5 5 0.184 0.820
46 0.85242 False 6 3 0.195 0.767
47 0.86457 False 6 3 0.195 0.767
48 0.87673 False 6 3 0.195 0.767
49 0.88888 False 6 3 0.195 0.767
50 0.90104 True 6 1 0.206 0.739
51 0.91319 False 6 1 0.206 0.739
52 0.92534 False 6 1 0.206 0.739
53 0.93750 False 6 1 0.206 0.739
54 0.94965 False 6 1 0.206 0.739
55 0.96181 False 6 1 0.206 0.739
56 0.97396 False 6 1 0.206 0.739
57 0.98612 False 6 1 0.206 0.739
58 0.99827 False 6 0 0.202 0.683
59 1.01042 False 6 0 0.202 0.683
60 1.02258 False 6 0 0.202 0.683
retention mean_size max_size
40 0.70 2.333 4.0
41 0.70 2.333 4.0
42 0.70 2.333 4.0
43 0.70 2.333 4.0
44 0.75 2.500 5.0
45 0.75 3.000 5.0
46 0.85 2.833 5.0
47 0.85 2.833 5.0
48 0.85 2.833 5.0
49 0.85 2.833 5.0
50 0.95 3.167 5.0
51 0.95 3.167 5.0
52 0.95 3.167 5.0
53 0.95 3.167 5.0
54 0.95 3.167 5.0
55 0.95 3.167 5.0
56 0.95 3.167 5.0
57 0.95 3.167 5.0
58 1.00 3.333 5.0
59 1.00 3.333 5.0
60 1.00 3.333 5.0
The output contains up to 5 files when optimising, or 3 files when using a custom threshold:
Silhouette score, purity, retention are plotted in range (0-1) on the main Y-axis. mean size and n_cluster above are plotted in log2 scale on the secondary Y-axis. n_above denotes the number of clusters above size 6 and purity 80% (only useful if labels were provided)
Here, you will find the data set used for training and evaluating TCRcluster-1.0.
All partitions, binders only (Used to train models)
All partitions, with swapped negatives (Used to train models)
17 peptides subset, test set binders only (used for model development)
17 peptides subset, with swapped negatives (used for model development)
17 peptides subset, test set with swapped negatives (used for figure 2)
17 peptides subset, test set binders only (used for figure 3)
...
Submitted 2025.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: