DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
The NetMHCIIpan-4.3 server predicts peptide binding to HLA class II molecules using Artificial Neural Networks (ANNs). It is trained on an extensive dataset of over 650,000 measurements of Binding Affinity (BA) and Eluted Ligand mass spectrometry (EL), covering the three human MHC class II isotypes HLA-DR, HLA-DQ, HLA-DP, as well as mouse (H-2) and bovine (BoLA-DRB3) molecules.
The network can predict for any HLA class II molecule of known sequence, which the user can specify as FASTA format, and predictions can be made for peptides of any length.
The output of the model is a prediction score for the likelihood of a peptide to be naturally presented by an MHC-II receptor of choice. The output also includes a %rank score, which normalizes the prediction score by comparing to predictions of a set of random peptides. Optionally, the model also outputs BA prediction and %rank scores.
New in version 4.3: The method is trained on an extended EL dataset including new data for HLA-DP, HLA-DR and BoLA-DRB3. Further, the method allows for prediction of inverted peptide binders.
Refer to the instructions page for more details.
The project is a collaboration between DTU-Bioinformatics, and LIAI.
View the version history of this server
Updated 18 June 2025: Optimized version of the back-end code gives ~2X faster predictions. Downloadable package updated (version 4.3i).
For publication of results, please cite:
NetMHCIIpan 4.3 is available as a stand-alone software package, with the same functionality as the service above. Ready-to-ship packages exist for Linux and macOS. There is a download page for academic users; other users are requested to contact Health Tech Software Package Manager at health-software@dtu.dk.
In this section, the user must define the input for the prediction server following these steps:
1) Specify the desired type of input data (FASTA or PEPTIDE) using the drop down menu.
2) Provide the input data by pasting it into the blank field, uploading it using the "Choose File" button, or by loading sample data using the "Load Data" button. All input sequences must be in one-letter amino acid code. The alphabet is as follows (case sensitive):
A C D E F G H I K L M N P Q R S T V W Y and X (unknown)
Any other symbol will be converted to X before processing. A maximum of 5000 sequences are allowed per submission; each sequence must be between 9 and 20,000 amino acids long.
3) If FASTA was selected, the user must select the peptide length(s) for the prediction server. NetMHCIIpan-4.3 will "chop" the FASTA sequence into overlapping peptides of the selected length and predict binding for each. By default, input proteins are digested into 15-mer peptides. If PEPTIDE was selected, this step is unnecessary, and the peptide length selector will not appear.
4) Context encoding informs the network of the proteolytic context of the ligand. Context is automatically generated from the source protein if the user selects FASTA format. The context consists of 12 amino acids: 3 upstream of the ligand, 3 from the N-terminus, 3 from the C-terminus, and 3 downstream from the ligand. If PEPTIDE is selected, the user must specify the ligand context (see PEPTIDECONT).

In this section, the user must define which MHC molecule(s) to predict against:
1) Select MHC molecules from a list by selecting a group and choosing MHCs. MS-COVERED refers to molecules covered by the NetMHCIIpan-4.3 training data.
2) Alternatively, the user can type the molecule names. Both ALPHA and BETA chains must be typed (see List of MHC molecule names). Selections from step 1 populate this bar.
3) If the desired molecule is not in the list, the user can input ALPHA and BETA sequences in FASTA format. Rank score predictions are not available in this case, unless the pseudo-sequence generated from the full-length ALPHA and BETA sequences is among the method's pre-calculated score threshold files.

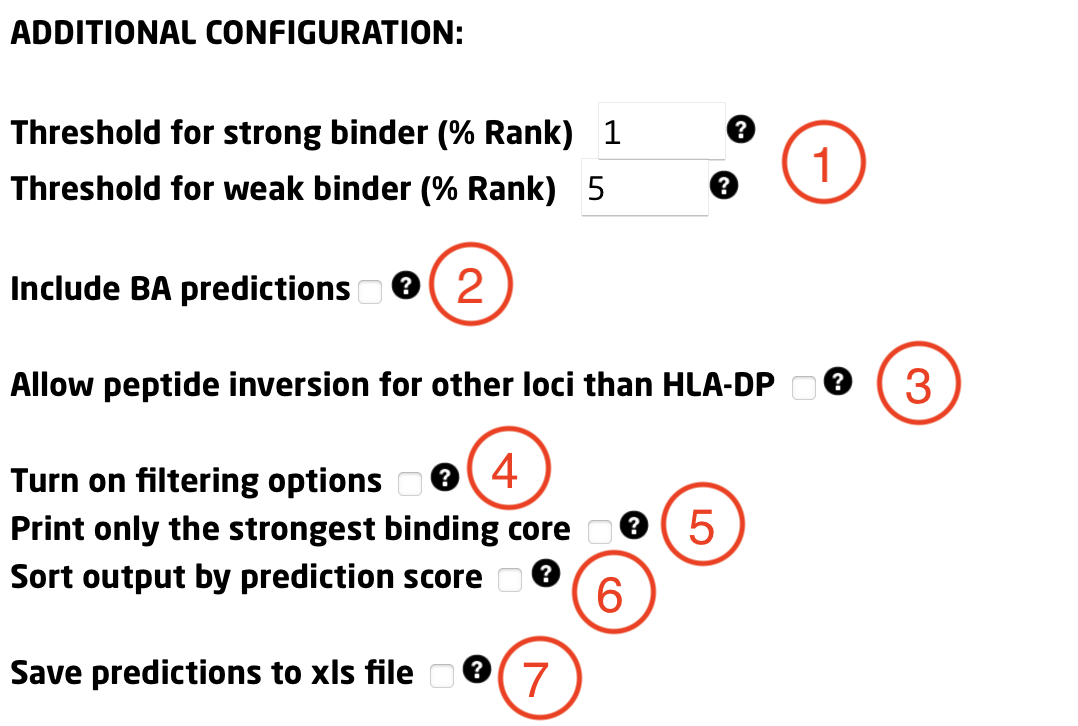
In this section, additional parameters can be defined to customize the run:
1) Specify thresholds for strong and weak binders (%Rank). Peptides identified in the top x% are strong binders (default: 1%). Peptides between strong and weak thresholds are weak binders (default: 5%).
2) Include Binding Affinity predictions alongside Eluted Ligand likelihood.
3) Enable peptide inversion prediction for all selected MHC-II molecules (optional; default is HLA-DP only).
4) Output only peptides below a specified %Rank score (useful for large submissions).
5) Output only the strongest binding core.
6) Sort output by descending prediction score.
7) Export output to .XLS format.
After completing the "INPUT DATA", "MHC SELECTION", and "ADDITIONAL CONFIGURATION" steps, the submission can now be done. Click "Submit" to send the job to the server, or click "Clear fields" to reset the form.
Job status ('queued' or 'running') will be displayed and updated until it terminates, and the output will appear in the browser window.
After completion, an output page will be delivered. A description of the output format can be found here.
You can enter your email address at any time to receive notification when the job is complete.

# NetMHCIIpan version 4.3i # Input is in FASTA format # Peptide length 15 # Prediction Mode: EL # Threshold for Strong binding peptides (%Rank) 1.00% # Threshold for Weak binding peptides (%Rank) 5.00% # HLA-DPA10202-DPB11901 : Distance to training data 0.000 (using nearest neighbor HLA-DPA10202-DPB11901) # Allele: HLA-DPA10202-DPB11901 -------------------------------------------------------------------------------------------------------------------------------------------- Pos MHC Peptide Of Core Core_Rel Inverted Identity Score_EL %Rank_EL Exp_Bind BindLevel -------------------------------------------------------------------------------------------------------------------------------------------- 25 HLA-DPA10202-DPB11901 NPISEFVKWYKSHKL 4 KYWKVFESI 0.980 1 Q5QFB9 0.600852 0.84 NA <= SB 24 HLA-DPA10202-DPB11901 WNPISEFVKWYKSHK 3 KYWKVFESI 0.970 1 Q5QFB9 0.567158 1.02 NA <= WB 26 HLA-DPA10202-DPB11901 PISEFVKWYKSHKLS 5 KYWKVFESI 0.930 1 Q5QFB9 0.459227 1.76 NA <= WB 23 HLA-DPA10202-DPB11901 IWNPISEFVKWYKSH 2 KYWKVFESI 0.880 1 Q5QFB9 0.319686 3.48 NA <= WB 27 HLA-DPA10202-DPB11901 ISEFVKWYKSHKLSQ 3 KHSKYWKVF 0.420 1 Q5QFB9 0.239845 5.02 NA 28 HLA-DPA10202-DPB11901 SEFVKWYKSHKLSQH 4 KWYKSHKLS 0.680 0 Q5QFB9 0.216487 5.63 NA 6 HLA-DPA10202-DPB11901 VRKKHRGLFLTTVAA 3 KHRGLFLTT 0.980 0 Q5QFB9 0.161751 7.49 NA 22 HLA-DPA10202-DPB11901 PIWNPISEFVKWYKS 1 KYWKVFESI 0.590 1 Q5QFB9 0.147204 8.15 NA 29 HLA-DPA10202-DPB11901 EFVKWYKSHKLSQHC 5 YKSHKLSQH 0.440 0 Q5QFB9 0.128036 9.16 NA 5 HLA-DPA10202-DPB11901 FVRKKHRGLFLTTVA 4 KHRGLFLTT 0.850 0 Q5QFB9 0.113368 10.07 NA
The prediction output for each molecule consists of the following columns:
Jonas B. Nilsson, Saghar Kaabinejadian, Hooman Yari, Michel G. D. Kester, Peter van Balen, William H. Hildebrand and Morten Nielsen
Science Advances, 24 Nov 2023. https://www.science.org/doi/10.1126/sciadv.adj6367Here, you will find the data set used for training of NetMHCIIpan-4.3.
Download the file and untar the content using
cat NetMHCIIpan_train.tar.gz | tar xvf -
This will create the directory called NetMHCIIpan_train. In this directory you will find 12 files. 10 files (c00?_ba, c00?_el) with partitions with binding affinity (ba) or eluted ligand data (el). The format for each file is (here shown for an el file)
AAAAMAEQESARN 1 Saghar_9061_DR MAAAAAARNGGR AAAAVQGGRSGG 1 Saghar_9090_DR MAAAAVSGGSGG AAALEAMKDYTKAM 1 Saghar_9013_DR TRKAAAKAMDVY AAALEAMKDYTKAMD 1 Saghar_9013_DR TRKAAAAMDVYQ AAEFIQQFNNQAFS 1 Saghar_9090_DR DKMAAEAFSVGQ AAEFIQQFNNQAFSVG 1 Saghar_9090_DR DKMAAESVGQQL AAFPFLAYSGIPAVS 1 Saghar_9013_DR LDNAAFAVSFCF AAGQFFPEAAQVAYQ 1 Saghar_9090_DR DDDAAGAYQMWE AAGVTDGNEVAKA 1 Saghar_9061_DR VRGAAGAKAQQA AAIRKKLVIVGD 1 Saghar_9013_DR MAAIRKVGDGACwhere the different columns are peptide, target value, MHC_molecule/cell-line, and context. In cases where the 3rd columns is a cell-line ID, the MHC molecules expressed in the cell-line are listed in the allelelist file.
The allelelist file contains the information about alleles expressed in each cell line data set, and pseudosequence.2023.dat the MHC pseudo sequence for each MHC molecule.
>ID Epitope HLA Sequencewhere ID is the Uniprot identifier, Epitope is the epitope, HLA is the HLA molecule bound by the epitope, and Sequence is the source protein sequence in which the epitope is derived.
Please click on the version number to activate the corresponding server.
| 4.3 |
The current version (online since July 2023). New in this version:
|
| 4.2 |
(online since September 2022). New in this version:
|
| 4.1 |
(online since Sept 2021). New in this version:
|
| 4.0 |
(online since April 2020). New in this version:
|
| 3.2 |
(online since January 2018). New in this version:
|
| 3.1 |
(online since December 2014). New in this version:
|
| 3.0 |
(online since June 2013). New in this version:
|
| 2.1 |
(online since 6 June 2011). New in this version:
|
| 2.0 |
(online since 17 Nov 2010). New in this version:
|
| 1.1 |
(online since 15 April 2010). New in this version:
|
| 1.0 |
Original version (online version until April 15 2010):
Main publication:
|
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: