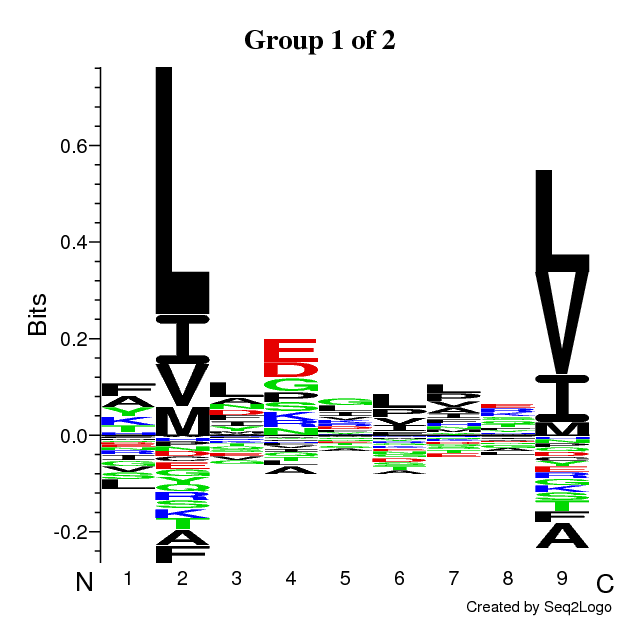
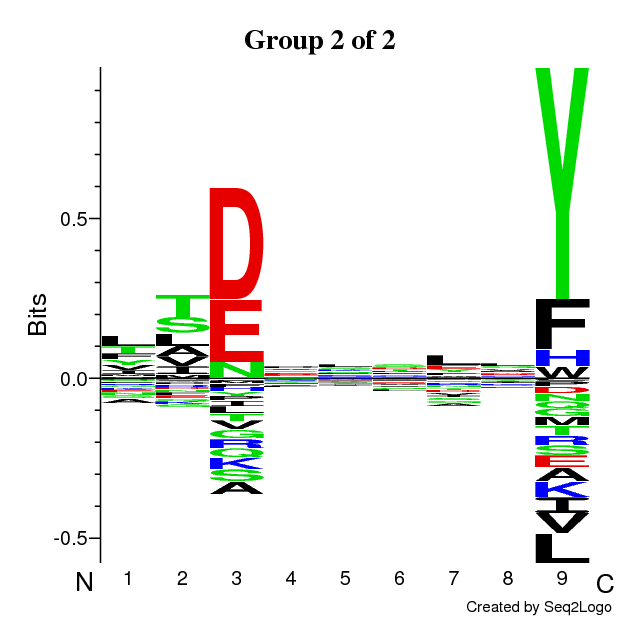
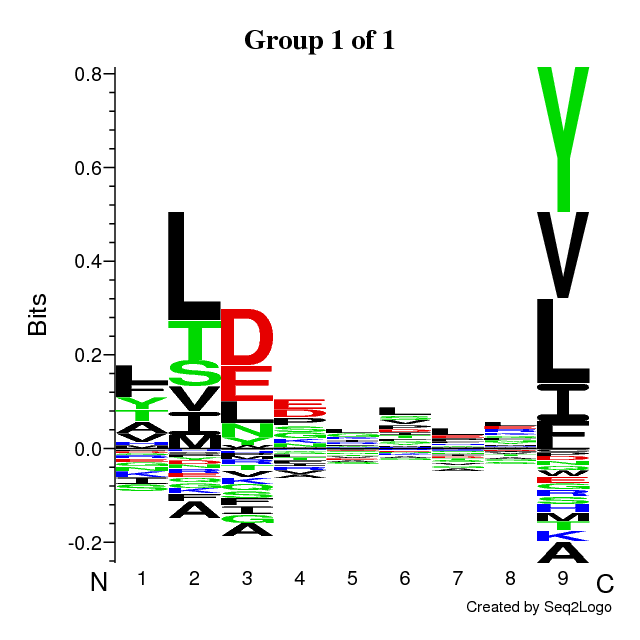
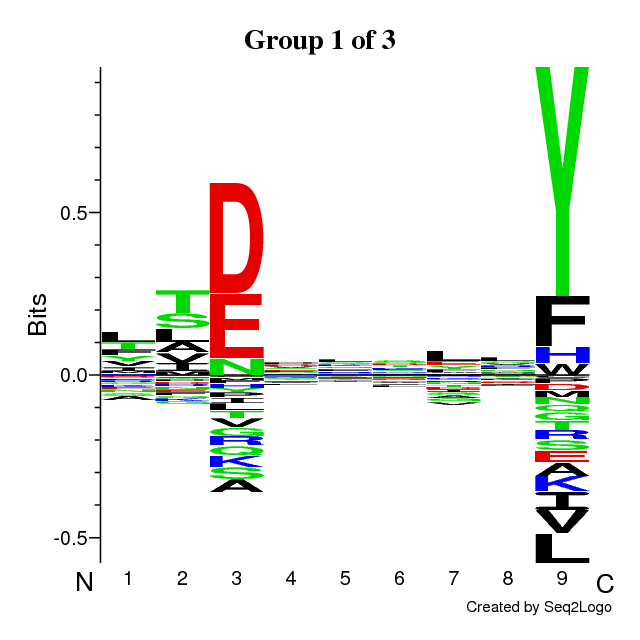
An example of output is found below. The output is composed of the following sections:
- Run identifiers and settings
The parameters specificied by the user are reported here, together with the number of
sequences loaded as input.
- Barplot of KLD vs number of clusters
For each initial number of clusters, the information content of the alignments is shown
as a barplot. The relative size of each block within a bar is proportional to the size of
a given cluster. In the example below, the run with 3 initial clusters produced one empty
cluster, therefore only 2 boxes are depicted in the third column of the barplot.
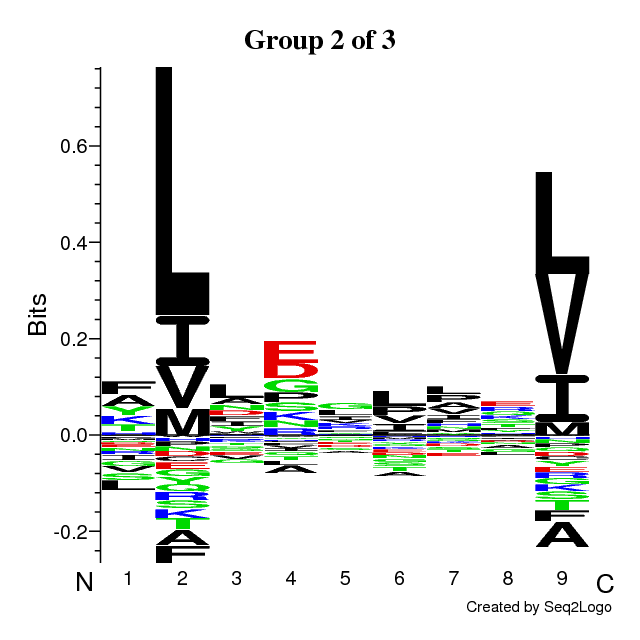
- Sequence logos of the optimal solution
The sequence motifs identified by the Gibbs Clustering are shown to the right of the
barplot. This is the optimal solution over the range of initial numbers of clusters.
Hovering the cursor over the logos shows the KLD of each cluster in the solution.
- Complete results for all initial number of clusters
If a range of initial numbers of clusters was specified (1 to 3 in the example below),
the results for each case are listed in succession. The optimal number of cluster shown
above is just a suggestion and depends on the specificied parameters, so the user is encouraged to inspect all solutions with different numbers of clusters.
It is possible to customize the logos further by clicking on the LOGO button.
This transfers the data to the Seq2Logo
server, which allows plotting several different kinds of sequence logo.
Inspect the complete Clustering Report and the formatted Clustering Solution
for a tabular version of the results. Format of the Clustering Solution files:
- Gn: Cluster number
- Num: Sequential number of the sequence in the cluster
- Sequence: Complete peptide sequence
- Core: Portion of the peptide in the alignment window
- of: Offset value, i.e. the starting position of the alignment core
- IP: Position of the insertion, if any
- IL: Length of the insertion, if any
- DP: Position of the deletion, if any
- DL: Length of the deletion, if any
- Annotation: Text annotation to the peptide, if provided at submission
- Self: Score of the peptide to its own cluster
- bgG: Identifier of the nearest cluster (-1 means no nearest cluster)
- bgScore: Score of the peptide to the nearest cluster
- cScore: corrected score (Sself - λ x Snearest)
Remember that results are only stored on the CBS server for about 24 hours.
Save your results to disk by clicking on the DOWNLOAD link at the bottom of the
results page.