Output format
DESCRIPTION
Example of output is found below. The output is divided into the folowinng sections:
- Description of training data
- Prediction method
- Parameters for training of Gibbs method
- Prediction data
- Evaluation of predictions (if assignments are supplied by user)
- Predictions
This section contain a line "Peptide Start res Motif Prediction (Assign) Sequence"
Peptide: Peptide number
Start res: 1st residue in motif
Motif: Motif found in sequence
Prediction: Prediction score for motif
Assign: Assignment of sequence (if supplied by user)
Sequence: Sequence containing motif
EXAMPLE OUTPUT
Description of training data
Length of motif: 9
Number of training data: 456
Number of positive training examples: 456
Parameters for training of Gibbs method
Clustering using the Henikoff & Henikoff 1/nr method
Weight on prior: 50.000000
Start temperature: 0.150000
End temperature: 0.000100
Number of temperature steps: 10.000000
Using default seed
Number of iterations per train example: 20
Using amino acid background distribution from SWISSPROT
Equal weights on all positions
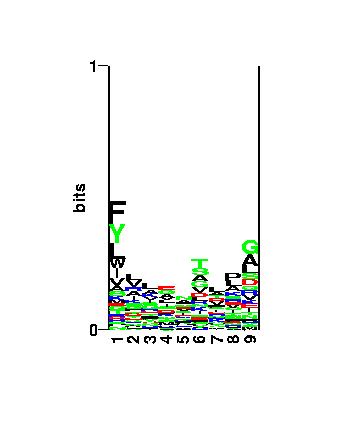
Figure: Visualization of the binding motif using the
logo
program.
A short explanation of HLA supertypes can be found
here.
Alignment generated by Gibbs sampler
Matrix generated by Gibbs sampler
Prediction data
Number of evaluation data: 57
Predicting using a matrix method
Evaluation of predictions
Pearson coefficient for N= 57 data: -0.30553
Aroc value: 0.31622
Threshold for counting example as positive: 50.000000
Predictions
Peptide Start res Motif Prediction Assign Sequence
1 8 FWSFGSEDG 6.549 5.400 MASPGSGFWSFGSEDGSGDS
2 1 FGSEDGSGD 3.304 62.000 FGSEDGSGDSENPGRARAWC
3 6 ARAWCQVAQ 3.363 100.000 ENPGRARAWCQVAQKFTGGI
4 1 QVAQKFTGG 1.638 100.000 QVAQKFTGGIGNKLCALLYG
5 8 LYGDAEKPA 4.603 100.000 GNKLCALLYGDAEKPAESGG
6 7 ESGGSQPPR 1.463 100.000 DAEKPAESGGSQPPRAAARK
7 8 ARKAACACD 3.170 100.000 SQPPRAAARKAACACDQKPC
8 12 CSKVDVNYA -1.160 2.400 AACACDQKPCSCSKVDVNYA
9 12 LHATDLLPA 6.138 0.500 SCSKVDVNYAFLHATDLLPA
10 2 LHATDLLPA 6.138 0.200 FLHATDLLPACDGERPTLAF
11 12 QDVMNILLQ 1.590 100.000 CDGERPTLAFLQDVMNILLQ
12 11 YVVKSFDRS 4.430 0.700 LQDVMNILLQYVVKSFDRST
13 1 YVVKSFDRS 4.430 19.000 YVVKSFDRSTKVIDFHYPNE
14 5 FHYPNELLQ 6.535 5.000 KVIDFHYPNELLQEYNWELA
15 7 WELADQPQN 7.249 5.000 LLQEYNWELADQPQNLEEIL
16 11 MHCQTTLKY 5.006 100.000 DQPQNLEEILMHCQTTLKYA
17 9 YAIKTGHPR 9.089 100.000 MHCQTTLKYAIKTGHPRYFN
18 8 YFNQLSTGL 5.320 0.500 IKTGHPRYFNQLSTGLDMVG
19 12 AADWLTSTA 1.734 1.400 QLSTGLDMVGLAADWLTSTA
20 5 WLTSTANTN 4.697 41.000 LAADWLTSTANTNMFTYEIA
21 5 FTYEIAPVF 3.031 85.000 NTNMFTYEIAPVFVLLEYVT
22 4 MREIIGWPG 7.542 48.000 LKKMREIIGWPGGSGDGIFS
23 9 FSPGGAISN 0.479 80.000 PGGSGDGIFSPGGAISNMYA
24 9 YAMMIARFK 3.854 100.000 PGGAISNMYAMMIARFKMFP
25 11 EVKEKGMAA 3.955 25.000 MMIARFKMFPEVKEKGMAAL
26 4 EKGMAALPR 4.336 40.000 EVKEKGMAALPRLIAFTSEH
27 6 FTSEHSHFS 8.330 0.200 PRLIAFTSEHSHFSLKKGAA
28 3 FSLKKGAAA 5.693 100.000 SHFSLKKGAAALGIGTDSVI
29 10 ILIKCDERG 1.002 24.000 ALGIGTDSVILIKCDERGKM
30 10 MIPSDLERR -0.681 100.000 LIKCDERGKMIPSDLERRIL
31 8 RILEAKQKG 2.013 38.000 IPSDLERRILEAKQKGFVPF
32 10 FLVSATAGT 5.278 4.000 EAKQKGFVPFLVSATAGTTV
33 11 YGAFDPLLA 6.422 7.000 LVSATAGTTVYGAFDPLLAV
34 1 YGAFDPLLA 6.422 100.000 YGAFDPLLAVADICKKYKIW
35 10 WMHVDAAWG 9.248 2.700 ADICKKYKIWMHVDAAWGGG
36 1 MHVDAAWGG 1.533 43.000 MHVDAAWGGGLLMSRKHKWK
37 9 WKLSGVERA 7.137 0.800 LLMSRKHKWKLSGVERANSV
38 9 SVTWNPHKM 4.403 13.000 LSGVERANSVTWNPHKMMGV
39 5 HKMMGVPLQ 3.161 34.000 TWNPHKMMGVPLQCSALLVR
40 8 LVREEGLMQ 5.674 17.000 PLQCSALLVREEGLMQNCNQ
41 3 GLMQNCNQM 2.757 41.000 EEGLMQNCNQMHASYLFQQD
42 5 YLFQQDKHY 5.432 22.000 MHASYLFQQDKHYDLSYDTG
43 5 LSYDTGDKA 4.032 31.000 KHYDLSYDTGDKALQCGRHV
44 12 VFKLWLMWR 0.431 100.000 DKALQCGRHVDVFKLWLMWR
45 5 LWLMWRAKG 8.337 33.000 DVFKLWLMWRAKGTTGFEAH
46 7 FEAHVDKCL 2.003 100.000 AKGTTGFEAHVDKCLELAEY
47 12 YNIIKNREG 3.638 34.000 VDKCLELAEYLYNIIKNREG
48 2 YNIIKNREG 3.638 4.000 LYNIIKNREGYEMVFDGKPQ
49 2 EMVFDGKPQ 2.819 67.000 YEMVFDGKPQHTNVCFWYIP
50 7 WYIPPSLRT 5.691 0.600 HTNVCFWYIPPSLRTLEDNE
51 3 LRTLEDNEE 0.154 5.000 PSLRTLEDNEERMSRLSKVA
52 4 SRLSKVAPV 3.410 100.000 ERMSRLSKVAPVIKARMMEY
53 10 YGTTMVSYQ 2.533 10.000 PVIKARMMEYGTTMVSYQPL
54 3 TMVSYQPLG 1.012 10.000 GTTMVSYQPLGDKVNFFRMV
55 7 FRMVISNPA 11.762 0.700 GDKVNFFRMVISNPAATHQD
56 2 SNPAATHQD 3.258 65.000 ISNPAATHQDIDFLIEEIER
57 8 FLIEEIERL 3.306 20.000 ATHQDIDFLIEEIERLGQDL
