DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
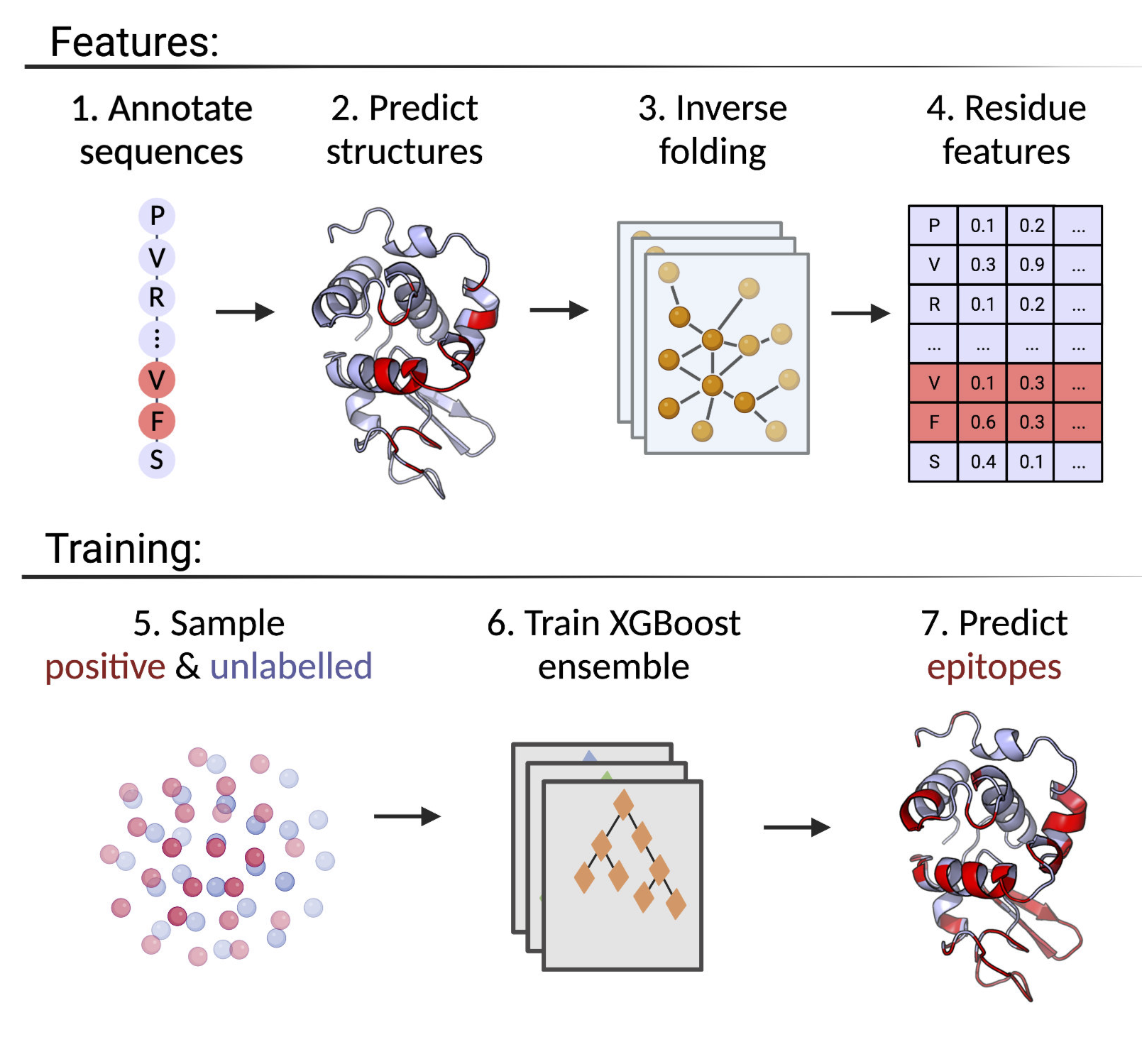
The DiscoTope server predicts B-cell epitopes from protein three dimensional structures. DiscoTope version 3.0 uses improved B-cell epitope prediction using AlphaFold2 modeling and inverse folding latent representations.
Mirror Use DiscoTope-3.0 on BioLib if this server is heavily loaded.
Upload or provide a list of PDB protein structures. For detailed instructions, see the Instructions tab.
DiscoTope-3.0 predicts per-residue epitope propensity of input protein structures. The server requires input protein structures in the PDB format. These may either be uploaded by the user or downloaded by the DiscoTope-3.0 server.
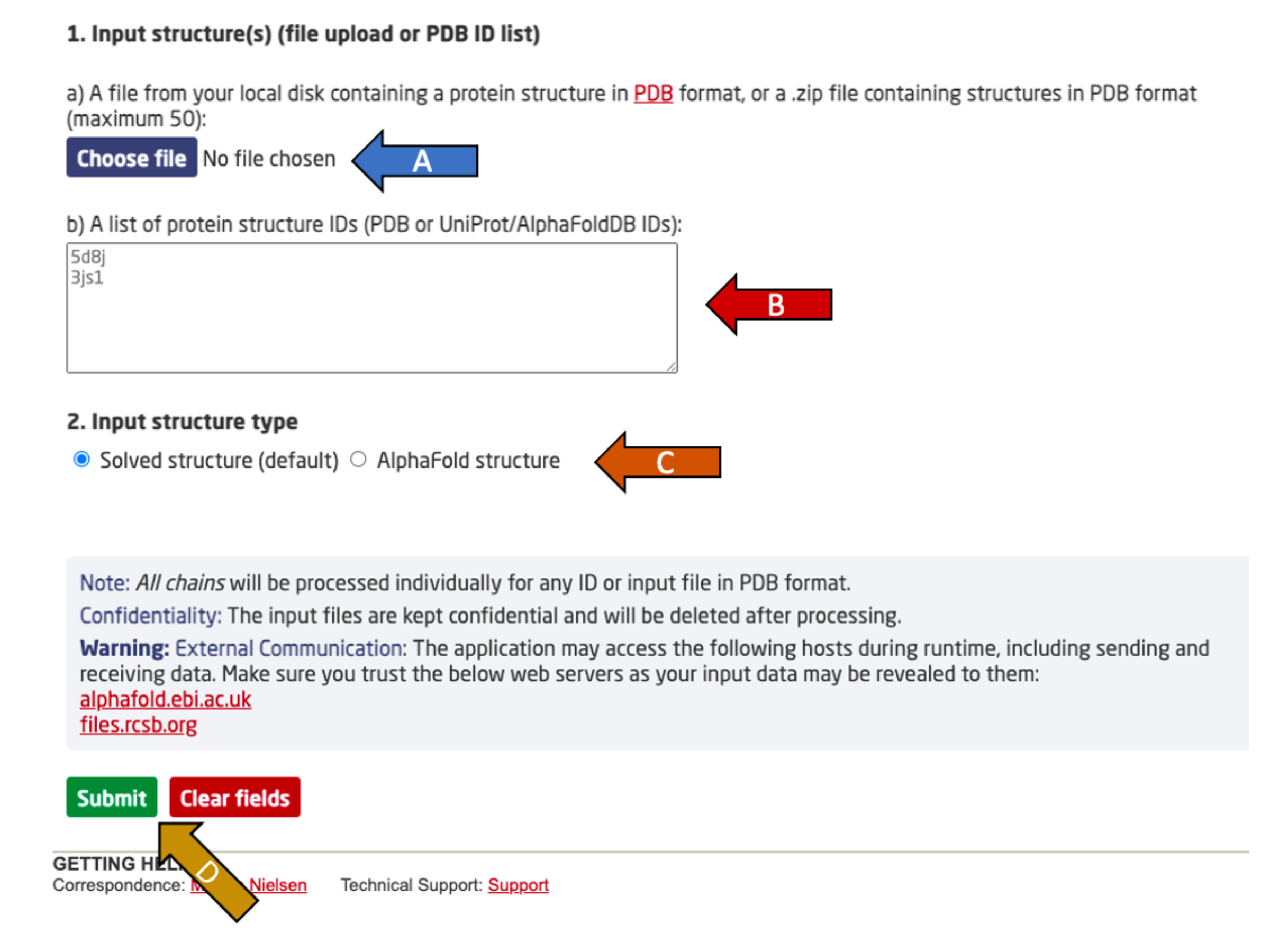
Choose between either:
After providing one of the two input options, specify the input structure type (arrow C). Experimental PDB structures (solved) and AlphaFold2 predicted structures are supported.
The suggested calibrated score thresholds correspond to observed epitope percentile scores in the validation set, matching expected recall rates at the given thresholds (see paper).
Finally, click submit to complete the submission (arrow D).
DiscoTope-3.0 outputs single chain PDB files, with matching per-residue CSV files. These can be visualized on the server with the Mol* viewer.
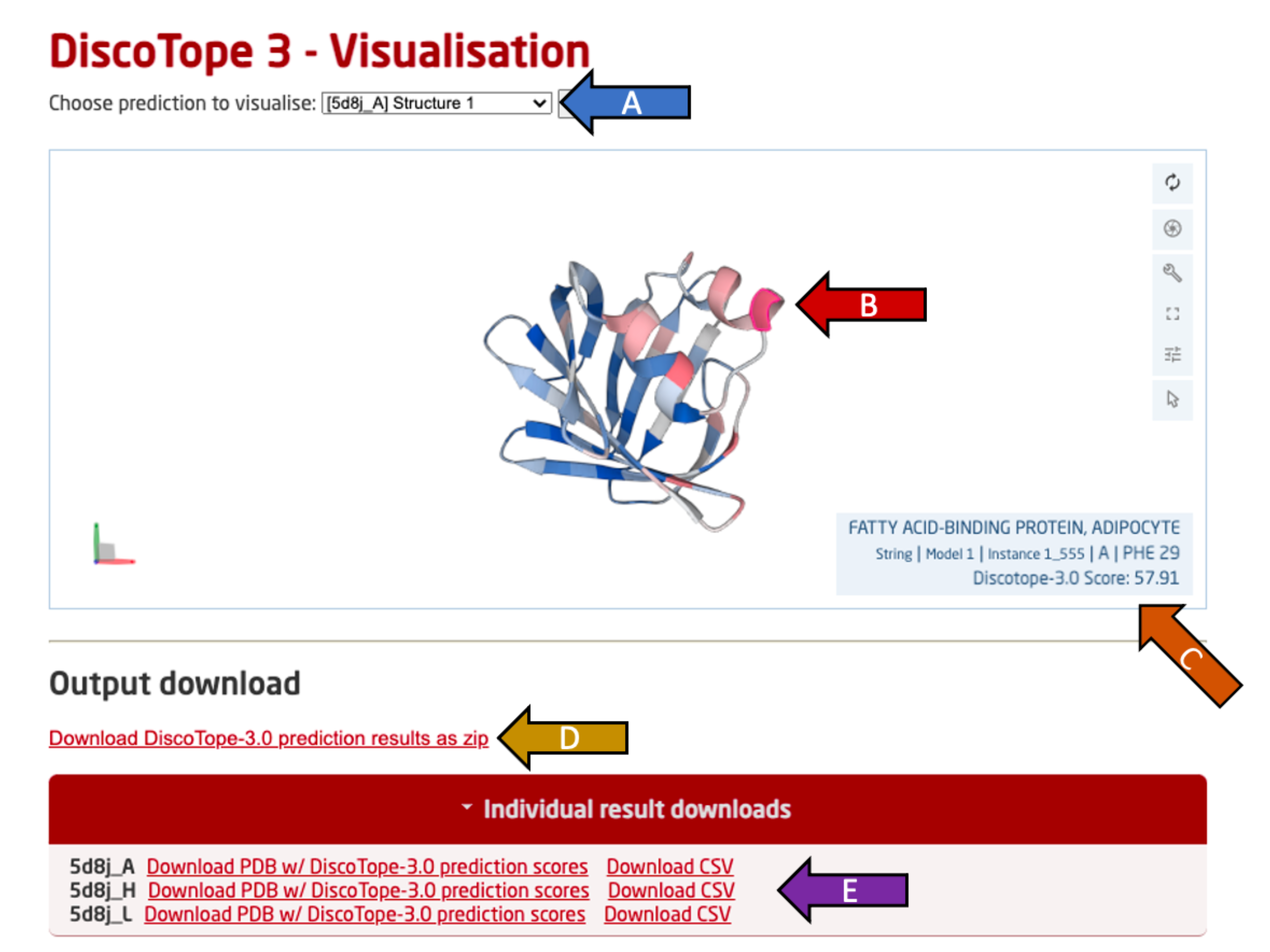
Use the drop-down menu (arrow A) to switch the view to another predicted PDB chain among all provided PDB files.
The selected chain is visualized in an interactive view below using Mol* (arrow B). This view may be rotated freely (click and move), along an axis (hold shift), zoomed (scroll) or moved (hold ctrl) using the mouse and listed keys in parantheses.
Move the cursor over a residue to see it's predicted DiscoTope-3.0 score, residue ID as well as the chain name (arrow C). Residues with higher epitope propensity are colored in a deeper red, while residues with lower epitope propensity are colored in a deeper blue. The color scale is absolute and not adjusted per PDB.
All output predictions may be downloaded as a compressed ZIP archive (arrow D), containing per PDB chain predictions in both .CSV and .PDB format.
Single PDB chain results may be downloaded in a .CSV or .PDB format by clicking the "Individual result downloads" button (arrow E).
The CSV files contains per-residue outputs, with the following column headers:The calibrated score thresholds correspond to observed epitope percentile scores in the validation set, matching expected recall rates at the given thresholds (see paper).
Mol* Viewer: modern web app for 3D visualization and analysis of large biomolecular structures
David Sehnal, Sebastian Bittrich, Mandar Deshpande, Radka Svobodová, Karel Berka, Václav Bazgier, Sameer Velankar, Stephen K Burley, Jaroslav Koča, Alexander S Rose.
Nucleic Acids Research (2021). doi: 10.1093/nar/gkab314
DiscoTope-3.0: Improved B-cell epitope prediction using inverse folding latent representations
Magnus Haraldson Høie, Frederik Steensgaard Gade, Julie Maria Johansen, Charlotte Würtzen, Ole Winther, Morten Nielsen, Paolo Marcatili.
Frontiers in Immunology (Feb 2024). doi: 10.3389/fimmu.2024.1322712
Accurate computational identification of B-cell epitopes is crucial for the development of vaccines, therapies, and diagnostic tools. Structure-based prediction methods generally outperform sequence-based models, but are limited by the availability of experimentally solved structures. Here, we present DiscoTope-3.0, a B-cell epitope prediction tool that exploits inverse folding representations from solved or AlphaFold-predicted structures. On independent datasets, the method demonstrates improved performance on both linear and non-linear epitopes with respect to current state-of-the-art algorithms. Most notably, our tool maintains high predictive performance across solved and predicted structures, alleviating the need for experiments and extending the general applicability of the tool by more than 4 orders of magnitude. DiscoTope-3.0 is available as a web server and downloadable package, processing up to 50 structures per submission. The web server interfaces with RCSB and AlphaFoldDB, enabling large-scale prediction on all currently cataloged proteins. DiscoTope-3.0 is available here and on BioLib.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: