DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
 |
 |
Unavailable - currently non-functional, no ETA on updatesThe YinOYang WWW server produces neural network predictions for O-ß-GlcNAc attachment sites in eukaryotic protein sequences. This server can also use NetPhos, to mark possible phosphorylated sites and hence identify "Yin-Yang" sites. |
Sequence submission: paste the sequence(s) or upload a local file
Restrictions:
At most 2000 sequences and 200000 amino acids per submission;
each sequence not less than 21 and not more than 4000 amino acids.
Confidentiality:
The sequences are kept confidential and will be deleted
after processing.
For publication of results, please cite:
Prediction of glycosylation sites in proteomes: from post-translational
modifications to protein function.
R Gupta.
Ph.D. thesis at CBS, 2001.
Prediction of glycosylation across the human proteome and the correlation
to protein function.
Gupta, R. and S. Brunak.
Pacific Symposium on Biocomputing, 7:310-322, 2002.
PMID: 11928486 Download the full article in PDF.
The YinOYang server incorporates results from NetPhos and SignalP.
In order to use the YinOYang WWW server for prediction on amino acid sequences:
The sequence must be written using the one letter amino acid code:
`acdefghiklmnpqrstvwy' or
`ACDEFGHIKLMNPQRSTVWY'.
Other letters will be converted to `X' and treated as unknown
amino acids.
Other characters, such as whitespace and
numbers, will simply be ignored.
Name: HXA3_HUMAN Length: 443 (sequence) MQKATYYDSSAIYGGYPYQAANGFAYNANQQPYPASAALGADGEYHRPACSLQSPSSAGGHPKAHELSEACLRTLSAPPS 80 QPPSLGEPPLHPPPPQAAPPAPQPPQPAPQPPAPTPAAPPPPSSASPPQNASNNPTPANAAKSPLLNSPTVAKQIFPWMK 160 ESRQNTKQKTSSSSSGESCAGDKSPPGQASSKRARTAYTSAQLVELEKEFHFNRYLCRPRRVEMANLLNLTERQIKIWFQ 240 NRRMKYKKDQKGKGMLTSSGGQSPSRSPVPPGAGGYLNSMHSLVNSVPYEPQSPPPFSKPPQGTYGLPPASYPASLPSCA 320 PPPPPQKRYTAAGAGAGGTPDYDPHAHGLQGNGSYGTPHIQGSPVFVGGSYVEPMSNSGPALFGLTHLPHAASGAMDYGG 400 AGPLGSGHHHGPGPGEPHPTYTDLTGHHPSQGRIQEAPKLTHL (annotation line, G=O-GlcNAc, Y=YinYang) ...................................G...................GY..................G...G 80 ..................................Y.......GG.Y.....G...G........................ 160 .....Y...GYYY..........Y.....Y.....G..G......................................... 240 ................G.Y...Y.G...........................Y....G.....G......G...G..G.. 320 ........................................................................G....... 400 ........................................G..
------------------------------------------------------------------------------- SeqName Residue O-GlcNAc Potential Thresh. Thresh. NetPhos YinOYang? result (o-glcnac) (1) (2) potential (Thresh=0.5) ------------------------------------------------------------------------------- HXA3_HUMAN 5 T - 0.4047 0.4125 0.5064 <-- A negative site HXA3_HUMAN 9 S - 0.4267 0.4631 0.5746 HXA3_HUMAN 10 S - 0.3548 0.4653 0.5776 HXA3_HUMAN 36 S ++ 0.5781 0.4421 0.5463 <-- O-GlcNAc predicted (++) HXA3_HUMAN 51 S - 0.4089 0.4299 0.5298 HXA3_HUMAN 54 S - 0.3183 0.3966 0.4849 0.783 <- Phos,No-GlcNAc HXA3_HUMAN 56 S + 0.4483 0.3792 0.4615 HXA3_HUMAN 57 S* + 0.4357 0.3856 0.4702 0.967 * <- YinYang HXA3_HUMAN 68 S - 0.2631 0.4479 0.5542 HXA3_HUMAN 74 T - 0.3126 0.4422 0.5464 HXA3_HUMAN 76 S ++ 0.5023 0.3922 0.4791 HXA3_HUMAN 80 S +++ 0.6007 0.3667 0.4447 HXA3_HUMAN 84 S - 0.2640 0.3769 0.4584 0.991 HXA3_HUMAN 115 T* +++ 0.6261 0.3591 0.4344 0.770 * <- YinYang
. . . -------------------------------------------------------------------------------
O-GlcNAc Result
This can be one of 5 possibles:-
- No O-GlcNAc predicted
O-GlcNAc predicted: (different strengths)
+ Potential > Thresh-1
++ Potential > Thresh-2
(Thresh-2 is a threshold based on more stringent surface measures)
+++ Potential > (Thresh-2
+ 0.1)
++++ Potential > (Thresh-2
+ 0.1) AND Potential >= 0.75
NetPhos
These potentials are displayed if 'YinYang' output was selected and
if the NetPhos potential
crosses the NetPhos (fixed) threshold.
Predictions are run in parallel from the NetPhos
server.
YinOYang
Ser/Thr residues which are predicted to be O-GlcNAcylated as well as
phosphorylated are
marked by an asterisk (*) in both the residue and the YinOYang
column. Such sites may be
reversibly and dynamically modified by O-GlcNAc or Phosphate groups
at different times
in the cell.
SignalP
With all predictions, the SignalP
server is run in parallel. If a sequence is predicted to contain
a signal peptide, a warning is displayed. Such sequences are unlikely
to be intracellular, and hence
unlikely to be O-GlcNAcylated.
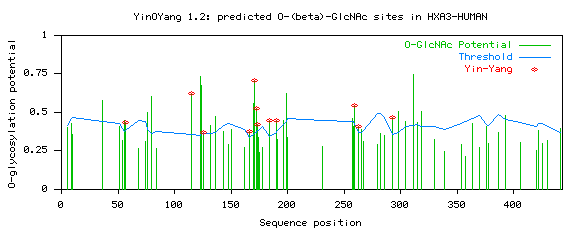
Graph
The figure illustrates O-GlcNAc and NetPhos predictions across the length
of the sequence.
The x-axis represents the sequence from N-terminal to C-terminal. Vertical
impulses (green)
are O-GlcNAc potentials. For predicted O-GlcNAcylated sites,
these potentials would cross
the threshold (blue wavy horizontal) line.
Small red marks/circles on
the green impulses indicate YinYang sites.
Intracellular O-glycosylation is characterised by the addition of N-acetylglucosamine, in a beta anomeric linkage, to Serine and Threonine residues in a protein. The acceptor site does not display a definite consensus sequence. However, the fuzzy motif is marked by the close vicinity of Proline residues (positions -4,-3,-2), Valines (-1,+2,+4,+5) and a downstream tract of Serines (+1,+4,+7) though Leucines and Glutamines are disfavoured. Secondary structure predictions indicate the 21-mer window to be sheet or coil. We train a jury of neural networks on 40 experimentally determined O-(beta)-GlcNAc acceptor sites, to recognise the sequence context and surface accessibility. Non-acceptor Serine/Threonines were pruned from 1251 in number to 626. In a cross-validation, 72.5% of the glycosylated sites and 79.5% of the non-glycosylated sites were correctly identified in the test set, revealing a Matthews correlation coefficient of 0.22 on the original data, and 0.84 on the augmented data set.
The method was used to scan all human protein sequences available in
SwissProt for potential O-(beta)-GlcNAc acceptors. Since this
modification is known to be reciprocal with phosphorylation, we
cross scanned for phosphorylation sites, and identified such
'Yin-Yang' sites. The spread of O-(beta)-GlcNAcylation, PEST regions
and phosphorylation sites, was studied across cellular role
categories, enzyme classes and subcellular compartments. Predicted
O-(beta)-GlcNAc sites were found in over half of all SwissProt human
sequences, 65% of which were nuclear or cytoplasmic.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: