DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
TatP 1.0 server predicts the presence and location of Twin-arginine signal peptide cleavage sites in bacteria. The method incorporates a prediction of cleavage sites and a signal peptide/non-signal peptide prediction based on a combination of two artificial neural networks. A postfiltering of the output based on regular expressions is possible.
| NOTE: TatP is outdated and is only kept online for reference. Tat signal peptides are better predicted by the current version of SignalP! |
For publication of results, please cite:
Prediction of twin-arginine signal peptides.
Jannick Dyrløv Bendtsen, Henrik Nielsen, David Widdick, Tracy Palmer
and Søren Brunak.
BMC bioinformatics 2005 6: 167.
All the alphabetic symbols not in the allowed alphabet will be converted to X before processing. All the non-alphabetic symbols, including white space and digits, will be ignored.
The sequences can be input in the following two ways:
Both ways can be employed at the same time: all the specified sequences will
be processed. However, there may be not more than 4,000 sequences and
2,000,000 amino acids in total in one submission. The sequences
may not be longer than 5,000 amino acids.
|
At any time during the wait you may enter your e-mail address and simply leave the window. Your job will continue; you will be notified by e-mail when it has terminated. The e-mail message will contain the URL under which the results are stored; they will remain on the server for 24 hours for you to collect them.
Description of the scores
The scores and graphical output is almost identical to the output
of the SignalP server. The presented scores are calculated in the
same way as for SignalP.
The graphical output from TatP (neural network) comprises three different
scores, C, S and Y. Two additional scores are
reported in the SignalP3-NN output, namely the S-mean and the
D-score, but these are only reported as numerical values.
For each prediction, two different neural networks are used, one for
predicting the actual signal peptide and one for predicting the
position of the signal peptidase I (SPase I) cleavage site. The
S-score for the signal peptide prediction is reported for
every single amino acid position in the submitted sequence, with
high scores indicating that the corresponding amino acid is part
of a signal peptide, and low scores indicating that the amino acid
is part of a mature protein.
The C-score is the ``cleavage site'' score. For each
position in the submitted sequence, a C-score is reported, which
should only be significantly high at the cleavage site. Confusion
is often seen with the position numbering of the cleavage site.
When a cleavage site position is referred to by a single number,
the number indicates the first residue in the mature protein,
meaning that a reported cleavage site between amino acid 26-27
corresponds to that the mature protein starts at (and include)
position 27.
Y-max is a derivative of the C-score combined with the
S-score resulting in a better cleavage site prediction than the
raw C-score alone. This is due to the fact that multiple
high-peaking C-scores can be found in one sequence, where only one
is the true cleavage site. The cleavage site is assigned from the
Y-score where the slope of the S-score is steep and a significant
C-score is found.
The S-mean is the average of the S-score, ranging from the
N-terminal amino acid to the amino acid assigned with the highest
Y-max score, thus the S-mean score is calculated for the length of
the predicted signal peptide.
The D-score is introduced in SignalP version 3.0 and is a
simple average of the S-mean and Y-max score. The score shows
superior discrimination performance of secretory and non-secretory
proteins to that of the S-mean score which was used in SignalP
version 1 and 2. In TatP the D-score is used for final discrimination
of secretory vs. non-secretory.
For non-secretory proteins all the scores represented in the
TatP-NN output should ideally be very low.
# data
Examples of standard output
Examples of short output
DESCRIPTION OF THE SCORES
EXAMPLES OF STANDARD OUTPUT
By default the server produces the following output for each input sequence:
Example 1: Secretory protein with Tat signal peptide
The example below shows the output for Membrane-bound hydrogenase 1 small subunit, taken from the
Swiss-Prot
entry MBHS_ECOLI.
The signal peptide prediction is consistent with the database annotation.
MBHS_ECOLI
TatP-NN result:
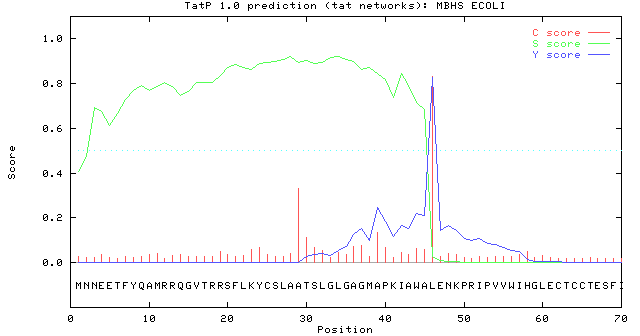
>MBHS_ECOLI length = 70
# Measure Position Value Cutoff signal peptide?
max. C 46 0.831 0.48 YES
max. Y 46 0.826 0.41 YES
max. S 34 0.923 0.84 YES
mean S 1-45 0.804 0.46 YES
max. D 1-45 0.815 0.44 YES
# Most likely cleavage site between pos. 45 and 46: AWA-LE
# Found RRQGV as Tat motif starting at position 12
Used regex: RR.[FGAVML][LITMVF]
//
EXAMPLE OF SHORT OUTPUT
When selecting the short output format, the prediction for each submitted
sequence (in a multisequence FASTA file) are reported in a condensed text form
without any graphical output. All entries are separated by a "//". The following
example show one positive and one negative prediction. The regular expression
entered on the webpage is also presented in the output.
>MBHS_ECOLI length = 70
# Measure Position Value Cutoff signal peptide?
max. C 46 0.831 0.48 YES
max. Y 46 0.826 0.41 YES
max. S 34 0.923 0.84 YES
mean S 1-45 0.804 0.46 YES
max. D 1-45 0.815 0.44 YES
# Most likely cleavage site between pos. 45 and 46: AWA-LE
# Found RRQGV as Tat motif starting at position 12
Used regex: RR.[FGAVML][LITMVF]
//
>AAT_THEMA length = 70
# Measure Position Value Cutoff signal peptide?
max. C 22 0.279 0.48 NO
max. Y 22 0.090 0.41 NO
max. S 6 0.102 0.84 NO
mean S 1-21 0.057 0.46 NO
max. D 1-21 0.073 0.44 NO
Used regex: RR.[FGAVML][LITMVF]
//
Abstract
Background:
Proteins carrying twin-arginine (Tat) signal peptides are exported into the periplasmic compartment or extracellular environment independently of the classical Sec-dependent translocation pathway. To complement other methods for classical signal peptide prediction we here present a publicly available method, TatP, for prediction of bacterial Tat signal peptides.
Results:
We have retrieved sequence data for Tat substrates in order to train a computational method for discrimination of Sec and Tat signal peptides. The TatP method is able to positively classify 91% of 35 known Tat signal peptides and 84% of the annotated cleavage sites of these Tat signal peptides were correctly predicted. This method generates far less false positive predictions on various datasets than using simple pattern matching. Moreover, on the same datasets TatP generates less false positive predictions than a complementary rule based prediction method.
Conclusions:
The method developed here is able to discriminate Tat signal peptides from cytoplasmic proteins carrying a similar motif, as well as from Sec signal peptides, with high accuracy. The method allows filtering of input sequences based on Perl syntax regular expressions, whereas hydrophobicity discrimination of Tat- and Sec-signal peptides is carried out by an artificial neural network. A potential cleavage site of the predicted Tat signal peptide is also reported.
PMID: 15992409 doi:10.1186/1471-2105-6-167
Upon public release of this method, more information will be added to this site.
Recently published papers regarding twin-arginine translocation can be found by quering PubMed.
A curated set of E. coli Tat signal peptides can be found at this website hosted by Tracy Palmer. E. coli dataset
Positive training set in fasta format
Download
Negative training set in fasta format Download
Negative test set (cytoplasmic RR)
Download
Output
Negative test set (transmem RR)
Download
Output
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: