Frequently Asked Questions
Changes from version 4 to 5
Changes from version 4.0 to 4.1
Changes from version 3 to 4
Biological background, signal peptides
Biological background, other sorting signals
Biological background, organism groups
History
— What's new?
Please see the version history page.
— What happened to the C-, S- and Y-scores?
The output layer of SignalP 5.0 is a conditional random field (CRF) which
yields marginal probabilities, just like the HMM module did in SignalP versions
2 and 3. Since the CRF is a grammatical method which is aware that there can
only be one cleavage site in a given signal peptide, there is no need for
the post-processing of the network output that was represented by the Y-score.
— What's new?
Please see the version history page.
— Why do you present a choice between two cutoff settings?
Can't you just decide on one?
The optimal cutoff really depends on what you want to use the method
for. If it is important to find all signal peptides, use the sensitive
cutoff. If you want an estimate of the number of signal peptides in a
genome, use the default cutoff.
— Why have you imposed a minimum length?
Because we believe that predictions of signal peptides
shorter than ten residues made by SignalP 4.1 are false. The shortest
known signal peptides are 11 residues long (with one exception,
SP23_TENMO,
which does not look like a signal peptide at
all). Click
here for an updated list of experimentally confirmed signal
peptides from UniProt of length 11 or shorter.
— What happened to the Background page?
It's here! The important material from the Background page has been
integrated into this FAQ, we hope you like the new format.
— What's new?
Please see the version history page.
— What happened to the HMM part?
While making SignalP 4.0, we did retrain the Hidden Markov Model (HMM)
part of SignalP. However, we found that it did not perform better than
the neural networks in any of the performance parameters we tested.
Therefore, we decided not to include it. If the HMM output is important
for you, you can still use
SignalP 3.0.
— Why is my favourite signal peptide no longer predicted correctly?
SignalP 3.0 could do it!
As explained on the performance page,
SignalP 4 with the default cutoff has a lower sensitivity than SignalP
3. Please try again with the new "Sensitive" setting.
— What happened to the Yes/No answers for max C score etc.?
SignalP 3.0 provided five Yes/No answers for the NN part. We found that
this was confusing for users and obscured the fact that the D-score is
the best score for discriminating between signal peptides and non-signal
peptides.
— What are signal peptides?
The term "signal peptide" is used with two meanings: In the broad
sense (used in many textbooks),
a signal peptide is any sorting signal embedded in the amino
acid sequence of a protein. In the narrow sense (used in most of
the scientific literature), a signal peptide
is an N-terminal signal that directs the protein across the ER
membrane in eukaryotes and across the plasma membrane in prokaryotes.
Signal peptides in the narrow sense are also known as ER signal
peptides or secretory signal peptides. Read more in
UniProt, in
Wikipedia,
and in the
Sequence feature ontology.
It is important to emphasize that SignalP predicts signal peptides in
the narrow sense only.
— Are signal peptides always N-terminal?
In the narrow sense: Yes, per definition. In the broad sense: No,
there are several sorting signal that are C-terminal
(e.g. the PTS1 signal for peroxisomal import)
or internal (e.g. the nuclear localization signal).
— Are signal peptides (in the narrow sense) always cleaved?
No, there are rare cases of uncleaved signal peptides. For an updated
list of such proteins annotated in UniProt, click
here.
These should not be confused with signal anchors, see below.
— Which protease is responsible for signal peptide (Sec/SPI)
cleavage?
In bacteria, it is Signal Peptidase I (SPase I), also known as Leader Peptidase
(Lep). In eukaryotes, it is the signal peptidase complex (SPC), which
consists of four subunits in yeast and five in mammals.
Read more in
MEROPS.
— My protein has a signal peptide. Can I then safely
conclude that it is secreted?
No. You can only conclude that it enters the secretory pathway.
In eukaryotes, there are several opportunities for a protein with a
signal peptide to escape secretion. It could:
- be retained in the endoplasmic reticulum (ER). Soluble ER-resident
proteins have a C-terminal retention signal with the consensus
sequence KDEL, see
PROSITE.
- be retained in the Golgi apparatus,
- be directed to the lysosome (vacuole in plants and fungi),
- have one or more transmembrane helices and therefore be
retained in either the plasma membrane, or one of the membranes of the
secretory pathway (ER, Golgi, lysosome/vacuole), or
- have a signal for GPI-anchoring, a C-terminal cleaved
peptide which functions as a signal for attachment of a
Glycophosphatidylinositol
(GPI) group that anchors the protein to the
outer face of the plasma membrane.
In Gram-positive bacteria and Archaea, a protein with a signal peptide could:
- have one or more transmembrane helices, or
- be attached to the cell wall.
In Gram-negative bacteria, a protein with a signal peptide could:
- have one or more transmembrane helices,
- be retained in the periplasm, or
- be inserted into the outer membrane as a β-barrel transmembrane
protein.
— Does SignalP predict signal peptides of bacterial and archaeal
lipoproteins?
Yes. Bacterial lipoproteins have special signal peptides (Sec/SPII) which are
cleaved by Signal Peptidase II (SPase II), also known as Lipoprotein
signal peptidase (Lsp). A diacylglyceryl group is attached to a Cysteine residue
in position +1 relative to the cleavage site, which bears no resemblance
to the SPase I cleavage site. See also
MEROPS
and PROSITE.
— Does SignalP predict Tat (Twin-arginine translocation) signal peptides?
Yes. Bacterial and archaeal Tat signal peptides (Tat/SPI), which direct their proteins through
an alternative translocon (TatABC instead of SecYEG),
have a special motif, usually containing two
Arginines, in the n-region. Additionally, they are in general longer and less
hydrophobic than "normal" (Sec) signal peptides. See also
PROSITE and
InterPro.
— What about Tat/SPII signal peptides?
SignalP cannot predict lipoprotein signal peptides that are transported through
the Tat translocon and cleaved by SPase II. When constructing the datasets
for SignalP 5.0, we gathered Tat signal peptides from
PROSITE PS51318 and
PRED-TAT,
while lipoprotein signal peptides were gathered from
PROSITE PS51257 and
PRED-LIPO, and there was
no overlap between them. even though such proteins are known to exist
(Thompson et al. 2010).
— What are signal anchors?
A signal anchor is a transmembrane helix located close to the N-terminus
of a protein with an N-in orientation (i.e. the N-terminus is on the
cytoplasmic side of the membrane). It functions much like a signal
peptide since it is recognized by the Signal Recognition Particle (SRP)
and inserted into the translocon; but instead of being cleaved and
degraded it remains in the membrane and anchors the protein to it.
Proteins anchored in this way are known as Type II transmembrane
proteins.
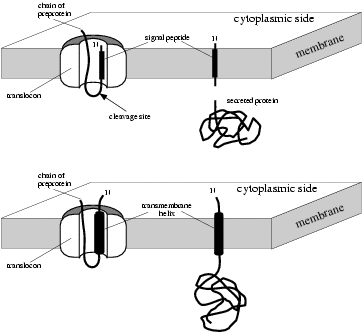 |
Signal peptides (above) versus
signal anchors (below) |
It is important to realize that the difference between signal peptides
and signal anchors is not a question of presence or absence of a
cleavage site. Instead, the most important difference seems to be the
length of the hydrophobic domain. It has been shown experimentally that
it is possible to convert a cleaved
signal peptide to a signal anchor merely by lengthening the
h-region, without altering the cleavage site
(Chou & Kendall 1990;
Nilsson, Whitley, & von Heijne 1994).
The introduction of the Hidden Markov Model (HMM) method in SignalP
version 2 made it possible to some extent to distinguish signal peptides
from signal anchors (in that version, only in eukaryotes). However,
SignalP 4 (based entirely on the Neural Network (NN) method), does a
better job, since its negative set is not confined only to transmembrane
helices annotated as signal anchors, but includes all types of
transmembrane segments close to the N-terminus.
— What should I use for predicting signal peptides in the
broad sense?
For mitochondrial and plastid import signals, also known as
transit
peptides, we recommend
TargetP. For
general prediction of subcellular location in eukaryotes, we recommend
DeepLoc.
— What should I use for predicting non-classical
(leaderless) secreted proteins?
Not all secretory proteins carry signal peptides. Some proteins enter a non-classical secretory pathway
without any currently known sequence motif. In eukaryotes, these proteins are mostly growth factors
and extracellular matrix binding proteins. In Gram-negative bacteria, the
type I, III, IV and VI secretion systems function without signal peptides.
For prediction of such proteins we
recommend the SecretomeP
server.
— Which version should I use for vira and bacteriophages?
You should use the version corresponding to the host organism. There are
some indications that viral signal peptides differ from those of the
host organism, but SignalP currently does not take that into account.
— Which version should I use for Tenericutes/Mollicutes
(Mycoplasma and related genera)?
You shouldn't use SignalP at all for these organisms, since they seem to
lack a type I signal peptidase completely!
— Which version should I use for metagenomic sequences
of unknown origin?
This is an unsolved question. Please use all four versions to
search for signal peptides in such data.
— Is one version enough for all eukaryotic organisms, or
are there differences within the eukaryotes?
It is known that some yeast signal peptides are not recognized by
mammalian cells (Bird et al.,
1987 and
1990).
Therefore, it would be natural to assume that separate SignalP versions
for yeast and Mammalia would provide better predictions than a common
eukaryotic version. While developing SignalP 4.0 we tried dividing the
eukaryotic data into animals, fungi, and plants and training separate
methods for these three groups. However, this did not give any
improvement, and performance for all three groups was better when using
the method trained on all eukaryotic sequences together.
— Are two versions enough for all bacteria, or
are there differences within the Gram-positive/Gram-negative
bacterial groups?
The Gram-negative version of SignalP is almost certainly biased towards
E. coli and other
γ-proteobacteria,
since these constitute the bulk
of the experimentally annotated bacterial proteins in UniProt.
Unpublished results suggest that some bacteria have very divergent
cleavage site motifs. Future versions of SignalP might therefore divide
the Gram-negative bacteria into several classes, if data are available.
Gram-positive bacteria probably constitute a more homogenous group, but
it is an open question whether there are differences in signal peptides
between
Actinobacteria (high G+C Gram-positive bacteria) and
Firmicutes (low G+C Gram-positive bacteria). More data on
Actinobacteria are needed before that can be answered.
— How are the various versions of SignalP related?
Please see the version history page
— Was there ever a Nobel prize awarded for signal peptides?
Yes, for signal peptides in the broad sense. The importance of signal peptides
was emphasized in 1999 when Günter Blobel received the Nobel Prize in
physiology or medicine for his discovery "proteins have intrinsic
signal that govern their transport and localization in the cell".
See the
press release.
— Was SignalP the first signal peptide predictor?
No, but it was, to our knowledge, the first to be implemented as a
web server (in 1996). Among the earlier methods were
McGeoch (1985)
and von Heijne (1986),
both of which have been included in
PSORT.
— How many times have the SignalP papers been cited?
This information is available on Henrik Nielsen's
ResearcherID,
Scopus,
and Google
Scholar pages.

