The best prediction of cleavage site location is provided by the position of the Y-score maximum. The best prediction of sequence type (signal peptide or non-secretory protein) is given by the mean S-score (the average of the S-score in the region between position 1 and the position immediately before the Y-score maximum): if mean S-score is larger than 0.5, the sequence is predicted to be a signal peptide (see the plot under ``Results: Identification of signal anchors''). When using these estimates, we obtain the predictive qualities given in the table below.
These prediction performances are minimal values. They are measured on the test sets (i.e. data which were not used to train the networks), and due to the redundancy reduction of the data, the sequence similarity between training and test sets is so low that the correct cleavage sites cannot be found by homology. Consequently, the prediction accuracy on sequences with some degree of homology to the sequences in the data sets will in general be higher.
| Version | Cleavage site location | Signal peptide discrimination | ||||
|---|---|---|---|---|---|---|
| EUK | Gram- | Gram+ | EUK | Gram- | Gram+ | |
| SignalP 1 NN | 70.2 | 79.3 | 67.9 | 0.97 | 0.88 | 0.96 |
| SignalP 2 NN | 72.44 | 83.43 | 67.46 | 0.97 | 0.90 | 0.96 |
| SignalP 2 HMM | 69.51 | 83.43 | 64.50 | 0.94 | 0.93 | 0.96 |
| SignalP 3 NN | 79.03 | 92.46 | 84.97 | 0.98 | 0.95 | 0.98 |
| SignalP 3 HMM | 75.70 | 90.22 | 81.58 | 0.94 | 0.94 | 0.98 |

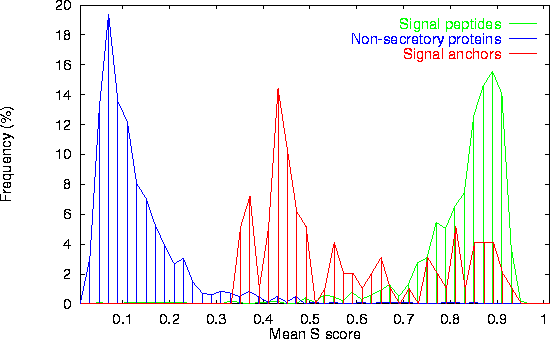
Above is shown the distribution of the mean S-score for three different protein types: Signal peptides, Non-secretory proteins (the N-terminal parts of cytoplasmic or nuclear proteins), and Signal anchors (the N-terminal parts of type II membrane proteins). Only eukaryotic data are shown here.
Signal anchors are also referred to as uncleaved signal peptides. However, they often have sites similar to signal peptide cleavage sites after their hydrophobic (transmembrane) region. Therefore, a prediction method can easily be expected to mistake signal anchors for peptides.
The mean S-score for signal anchors shows some overlap with the signal
peptide distribution (50% of the eukaryotic signal anchor sequences
have mean S-scores larger than 0.5). However, signal anchors are
generally significantly longer than signal peptides. By excluding
signal peptides longer than 35 residues (and using a slightly larger
cutoff), 72% of the eukaryotic signal anchor sequences are correctly
classified. (Only 2.2% of the cleaved eukaryotic signal peptides in
our data set are longer than 35 residues).