DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
| NOTE: SecretometP-2.0 is outdated and is only kept online for reference. We refer users to DeepLoc-2.1 and DeepLocPro-1.0 depending on your organism. |
Sequence submission: paste the sequence(s) and/or upload a local file
For publication of results, please cite as follows. When using predictions for mammalian sequences:
Feature based prediction of non-classical and leaderless protein secretion
J. Dyrløv Bendtsen, L. Juhl Jensen, N. Blom, G. von Heijne
and S. Brunak
Protein Eng. Des. Sel., 17(4):349-356, 2004
When using predictions for bacterial sequences:
Non-classical protein secretion in bacteria
J. Dyrløv Bendtsen, L. Kiemer, A. Fausbøll and S. Brunak
BMC Microbiology, 5:58, 2005
Please note that the sequences containing other symbols e.g. X (unknown) will be discarded before processing. The sequences can be input in the following two ways:
Both ways can be employed at the same time: all the specified sequences will
be processed. However, there may be not more than 100 sequences and 200,000 amino acids
in total in one submission. The sequences shorter than 15
or longer than 4000 amino acids will be ignored.
At any time during the wait you may enter your e-mail address and simply leave the window. Your job will continue; you will be notified by e-mail when it has terminated. The e-mail message will contain the URL under which the results are stored; they will remain on the server for 24 hours for you to collect them.
NOTE: SecretomeP is dependent on a number of other programs that have to be run on the input sequences prior to employing the SecretomeP method itself. Therefore, the processing of multiple sequences may be time-consuming. In the case of prolonged wait the user is advised to use the e-mail option mentioned above.
For eukaryotic sequences, an example of the output is shown below, but it is different in the version for bacteria. For that version, four scores are generated by the SecretomeP server for each input sequence. The determining score is the 'SecP score', for which a value above 0.5 indicates possible secretion. This score is the score mentioned in the reference paper, while the others are provided merely to give an idea about the degree of consent among the three different models used for the prediciton.
Even though SecretomeP is trained to predict non-classical secretion, it usually gives high score to proteins entering the classical secretory pathway (having signal peptides). The user is advised to check for conventional signal peptides if in doubt (See SignalP for more information).
# Name NN-score Odds Weighted Warning # by prior # ============================================================================ FGF1_HUMAN 0.847 4.267 0.009 - FGF4_HUMAN 0.945 6.804 0.014 signal peptide predicted by SignalP ATRX_HUMAN 0.093 0.205 0.000 -
Background: We present an overview of bacterial non-classical secretion and a prediction method for identification of proteins following signal peptide independent secretion pathways. We have compiled a list of proteins found extracellularly despite the absence of a signal peptide. Some of these proteins also have known roles in the cytoplasm, which means they could be so-called ``moon-lightning'' proteins having more than one function.
Methods: A thorough literature search was conducted to compile a list of currently known bacterial non-classically secreted proteins. Pattern finding methods were applied to the sequences in order to identify putative signal sequences or motifs responsible for their secretion. Finally, artificial neural networks were used to construct protein feature based methods for identification of non-classically secreted proteins in both Gram-positive and Gram-negative bacteria.
Results: We have found no signal or motif characteristic to any majority of the proteins in the compiled list of non-classically secreted proteins, and conclude that these proteins, indeed, seem to be secreted in a novel fashion. However, we also show that the apparently non-classically secreted proteins are still distinguished from cellular proteins by properties such as amino acid composition, secondary structure and disordered regions. Specifically, prediction of disorder reveals that bacterial secretory proteins are more structurally disordered than their cytoplasmic counterparts.
Conclusions: We present a publicly available prediction method capable of discriminating between this group of proteins and other proteins, thus allowing for the identification of novel non-classically secreted proteins. We suggest candidates for non-classically secreted proteins in Escherichia coli and Bacillus subtilis. The prediction method is available at http://www.cbs.dtu.dk/services/SecretomeP-2.0/.
PMID: 16212653
Gram-positives
Positive training set
Negative training set
Gram-negatives
Positive training set
Negative training set
Download file with top scoring proteins from E. coli excluding signal peptide containing sequences.
B. subtilis
Download file with top scoring proteins from B. subtilis excluding signal peptide containing sequences.
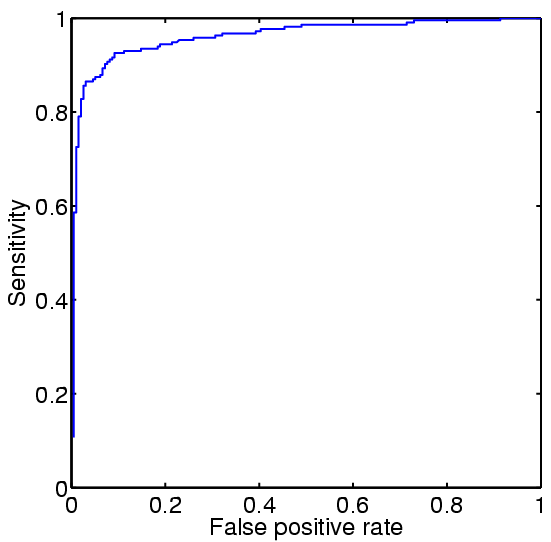
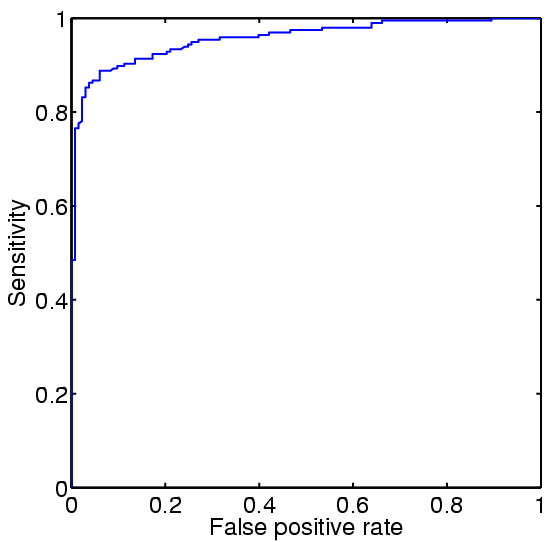
ROC curves for Gram-negative and Gram-positive, respectively.
Features for Gram-
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: