Instructions
1. Specify the input sequences
All the input sequences must be in one-letter amino acid
code. The allowed alphabet (not case sensitive) is as follows:
A C D E F G H I K L M N P Q R S T V W Y and X (unknown)
All the other symbols will be converted to X before processing. The
sequences can be input in the following two ways:
-
Paste a single sequence (just the amino acids) or a number of sequences in
FASTA
format into the upper window of the main server page.
-
Select a FASTA
file on your local disk, either by typing the file name into the lower window
or by browsing the disk.
Both ways can be employed at the same time: all the specified sequences will
be processed. However, there may be not more than 2,000 sequences and
200,000 amino acids in total in one submission. The sequences
may not be longer than 4,000 amino acids.
2. Customize your run
- By default the server produces graphical output illustrating
the predictions. You can suppress that by un-checking the button
labelled 'Generate graphics'.
- Prediction of the presence and location of signal peptide cleavage sites
by the SignalP server is included by
default. You can suppress that by un-checking the button labelled
'Include signal peptide prediction'.
- Check the button labelled 'Verbose output' to display the scores
produced by the 4 individual neural networks alongside the average score.
The default is to show the average score only.
- The server performs Furin-specific propeptide cleavage site prediction
by default. Check the button labelled 'General PC prediction'
to perform that prediction instead.
3. Submit the job
Click on the
"Submit" button. The status of your job (either 'queued'
or 'running') will be displayed and constantly updated until it terminates and
the server output appears in the browser window.
At any time during the wait you may enter your e-mail address and simply leave
the window. Your job will continue; you will be notified by e-mail when it has
terminated. The e-mail message will contain the URL under which the results are
stored; they will remain on the server for 24 hours for you to collect them.
Output format
DESCRIPTION
For each input sequence the following is shown (see the example below):
- Header line, quoting the name and length of the sequence.
- Sequence, in one-letter code, as it was interpreted by the server.
- Annotation overview, showing the predictions residue by residue,
as follows:
| . (dot) |
neutral place-holder |
| s |
predicted to be within a signal peptide |
| P |
predicted to be followed by a propeptide cleavage site |
- Prediction score table, quoting the position, context and score
for each arginine (R) and lysine (K) residue in the sequence.
If the score is >0.5 the residue is predicted to be followed by
a propeptide cleavage site; the higher the score the more confident
the prediction.
- Graph, in GIF, illustrating the predictions.
For each arginine (R) and lysine (K) the prediction score
is plotted against the position in the sequence. If a signal peptide
cleavage site has been predicted it is also shown in the graph.
The example below shows the output for
glial cell line-derived neurotrophic
factor precursor (ATF-1), taken from the
Swiss-Prot
entry
GDNF_HUMAN.
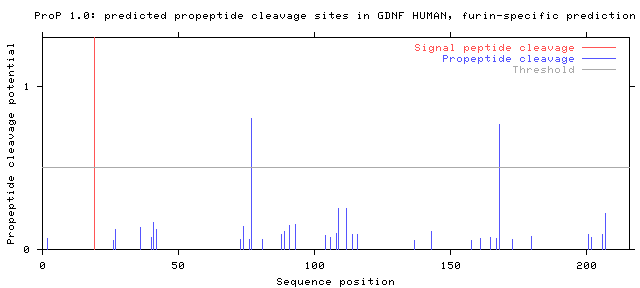
The signal peptide prediction is consistent with the database annotation.
Two propeptide cleavage sites are predicted; one of them (pos. 77-78) is
annotated in the database. The other prediction (pos. 168-169) is false.
EXAMPLE OUTPUT
##### ProP v.1.0b ProPeptide Cleavage Site Prediction #####
##### Furin-type cleavage site prediction (Arginine/Lysine residues) #####
211 GDNF_HUMAN
MKLWDVVAVCLVLLHTASAFPLPAGKRPPEAPAEDRSLGRRRAPFALSSDSNMPEDYPDQFDDVMDFIQATIKRLKRSPD 80
KQMAVLPRRERNRQAAAANPENSRGKGRRGQRGKNRGCVLTAIHLNVTDLGLGYETKEELIFRYCSGSCDAAETTYDKIL 160
KNLSRNRRLVSDKVGQACCRPIAFDDDLSFLDDNLVYHILRKHSAKRCGCI 240
sssssssssssssssssss.........................................................P... 80
................................................................................ 160
.......P........................................... 240
Signal peptide cleavage site predicted: between pos. 19 and 20: ASA-FP
Propeptide cleavage sites predicted: Arg(R)/Lys(K): 2
Name Pos Context Score Pred
____________________________v_________________
GDNF_HUMAN 2 -----MK|LW 0.069 .
GDNF_HUMAN 26 FPLPAGK|RP 0.054 .
GDNF_HUMAN 27 PLPAGKR|PP 0.120 .
GDNF_HUMAN 36 EAPAEDR|SL 0.134 .
GDNF_HUMAN 40 EDRSLGR|RR 0.074 .
GDNF_HUMAN 41 DRSLGRR|RA 0.163 .
GDNF_HUMAN 42 RSLGRRR|AP 0.124 .
GDNF_HUMAN 73 FIQATIK|RL 0.059 .
GDNF_HUMAN 74 IQATIKR|LK 0.140 .
GDNF_HUMAN 76 ATIKRLK|RS 0.062 .
GDNF_HUMAN 77 TIKRLKR|SP 0.802 *ProP*
GDNF_HUMAN 81 LKRSPDK|QM 0.062 .
GDNF_HUMAN 88 QMAVLPR|RE 0.090 .
GDNF_HUMAN 89 MAVLPRR|ER 0.108 .
GDNF_HUMAN 91 VLPRRER|NR 0.146 .
GDNF_HUMAN 93 PRRERNR|QA 0.152 .
GDNF_HUMAN 104 ANPENSR|GK 0.088 .
GDNF_HUMAN 106 PENSRGK|GR 0.071 .
GDNF_HUMAN 108 NSRGKGR|RG 0.099 .
GDNF_HUMAN 109 SRGKGRR|GQ 0.254 .
GDNF_HUMAN 112 KGRRGQR|GK 0.251 .
GDNF_HUMAN 114 RRGQRGK|NR 0.091 .
GDNF_HUMAN 116 GQRGKNR|GC 0.095 .
GDNF_HUMAN 137 GLGYETK|EE 0.058 .
GDNF_HUMAN 143 KEELIFR|YC 0.112 .
GDNF_HUMAN 158 AETTYDK|IL 0.054 .
GDNF_HUMAN 161 TYDKILK|NL 0.067 .
GDNF_HUMAN 165 ILKNLSR|NR 0.072 .
GDNF_HUMAN 167 KNLSRNR|RL 0.070 .
GDNF_HUMAN 168 NLSRNRR|LV 0.767 *ProP*
GDNF_HUMAN 173 RRLVSDK|VG 0.064 .
GDNF_HUMAN 180 VGQACCR|PI 0.080 .
GDNF_HUMAN 201 LVYHILR|KH 0.093 .
GDNF_HUMAN 202 VYHILRK|HS 0.074 .
GDNF_HUMAN 206 LRKHSAK|RC 0.095 .
GDNF_HUMAN 207 RKHSAKR|CG 0.222 .
____________________________^_________________
References
Prediction of proprotein convertase cleavage sites.
Peter Duckert, Søren Brunak and Nikolaj Blom1.
Protein Engineering, Design and Selection: 17: 107-112,
2004.
1to whom correspondence should be addressed, e-mail:
nikob@cbs.dtu.dk
Center for Biological Sequence Analysis, BioCentrum-DTU,
The Technical University of Denmark, DK-2800 Lyngby, Denmark
Abstract
Many secretory proteins and peptides are synthesized as inactive precursors
that in addition to signal peptide cleavage undergo post-translational
processing to become biologically active polypeptides. Precursors are
usually cleaved at sites composed of single or paired basic amino acid
residues by members of the subtilisin/kexin-like proprotein convertase (PC)
family. In mammals, seven members have been identified, with furin being the
one first discovered and best characterized. Recently, the involvement of
furin in diseases ranging from Alzheimer's disease and cancer to anthrax and
Ebola fever has created additional focus on proprotein processing. We have
developed a method for prediction of cleavage sites for PCs based on
artificial neural networks. Two different types of neural networks have been
constructed: a furin-specific network based on experimental results derived
from the literature, and a general PC-specific network trained on data from
the Swiss-Prot protein database. The method predicts cleavage sites in
independent sequences with a sensitivity of 95% for the furin neural network
and 62% for the general PC network.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: