Current version (NetSurfP-3.0 webserver)
NetSurfP-3.0: accurate and fast prediction of protein structural features by protein language models and deep learningMagnus Haraldson Høie, Erik Nikolas Kiehl, Bent Petersen, Henrik Nielsen, Ole Winther, Jeppe Hallgren, Paolo Marcatili.
Nucleic Acid Research (June 2022). doi: 10.1093/nar/gkac439
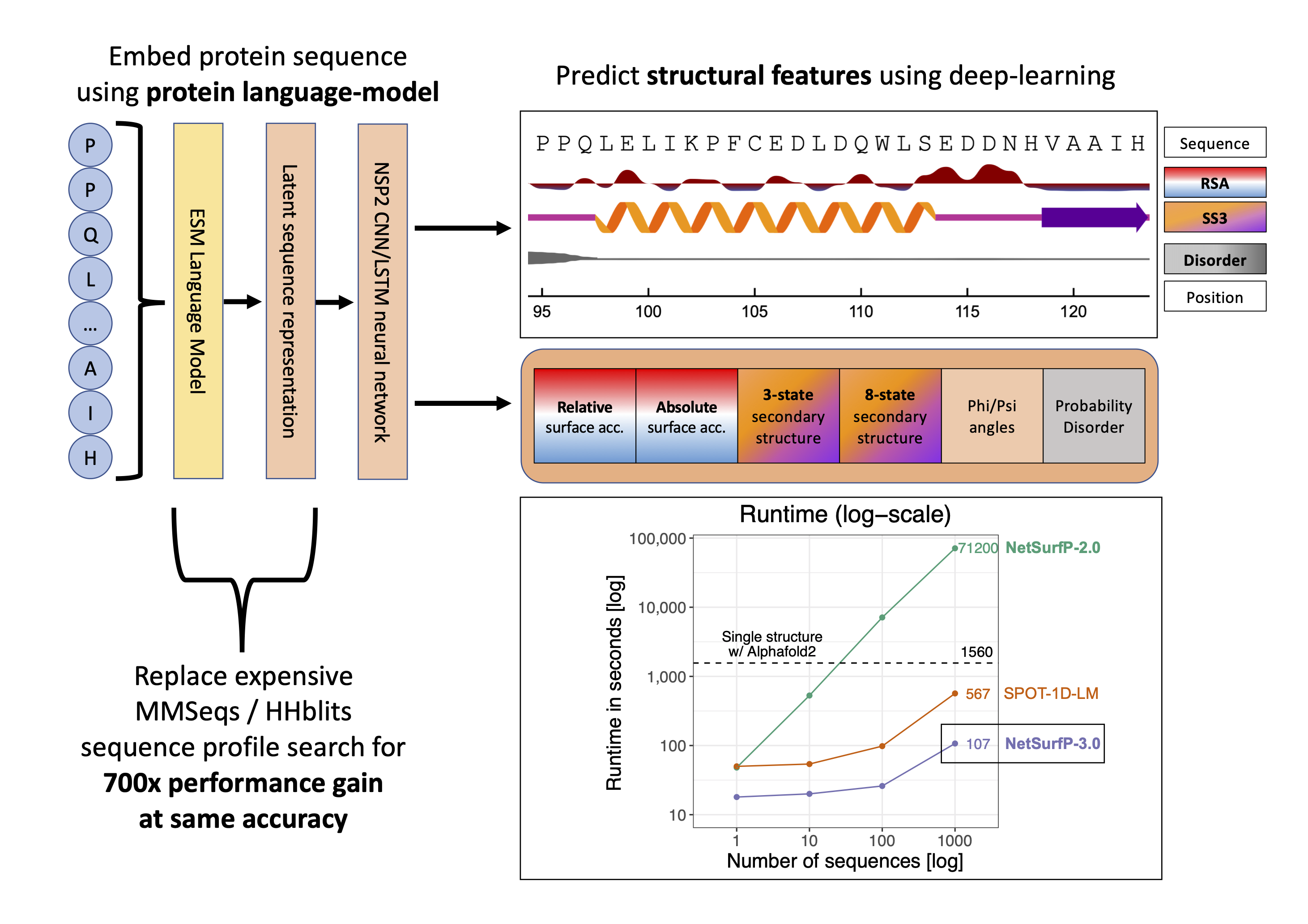
Recent advances in machine learning and natural language processing have made it possible to profoundly advance our ability to accurately predict protein structures and their functions. While such improvements are significantly impacting the fields of biology and biotechnology at large, such methods have the downside of high demands in terms of computing power and runtime, hampering their applicability to large datasets. Here, we present NetSurfP-3.0, a tool for predicting solvent accessibility, secondary structure, structural disorder and backbone dihedral angles for each residue of an amino acid sequence. This NetSurfP update exploits recent advances in pre-trained protein language models to drastically improve the runtime of its predecessor by two orders of magnitude, while displaying similar prediction performance. We assessed the accuracy of NetSurfP-3.0 on several independent test datasets and found it to consistently produce state-of-the-art predictions for each of its output features, with a runtime that is up to to 600 times faster than the most commonly available methods performing the same tasks. The tool is freely available as a web server with a user-friendly interface to navigate the results, as well as a standalone downloadable package.