Michael Schantz Klausen, Martin Closter Jespersen, Henrik Nielsen, Kamilla Kjærgaard Jensen, Vanessa Isabell Jurtz, Casper Kaae Sønderby, Morten Otto Alexander Sommer, Ole Winther, Morten Nielsen, Bent Petersen, and Paolo Marcatili. NetSurfP-2.0: Improved prediction of protein structural features by integrated deep learning . Proteins: Structure, Function, and Bioinformatics (Feb. 2019). doi: 10.1002/prot.25674
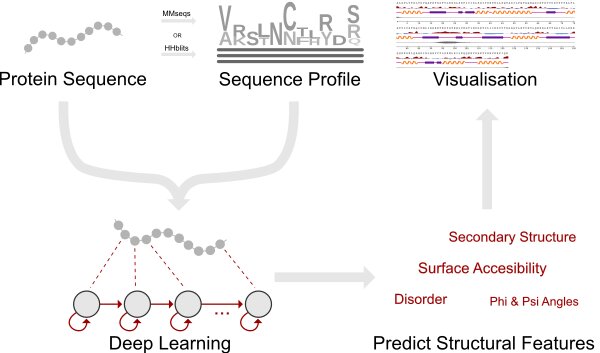
The ability to predict a protein’s local structural features from the primary sequence is of paramount importance for unraveling its function if no solved structures of the protein or its homologs are available. Here we present NetSurfP-2.0 ( https://services.healthtech.dtu.dk/service.php?NetSurfP-2.0 ), an updated and extended version of the tool that can predict the most important local structural features with unprecedented accuracy and run-time. NetSurfP-2.0 is sequence-based and uses an architecture composed of convolutional and long short-term memory neural networks trained on solved protein structures. Using a single integrated model, NetSurfP-2.0 predicts solvent accessibility, secondary structure, structural disorder, and backbone dihedral angles for each residue of the input sequences.
We assessed the accuracy of NetSurfP-2.0 on several independent validation datasets and found it to consistently produce state-of-the-art predictions for each of its output features, with a significant improvement for solvent accessibility and disorder. In addition to improved prediction accuracy the processing time has been optimized to allow proteome wide predictions of more than 4,000 proteins in less than 10 hours.