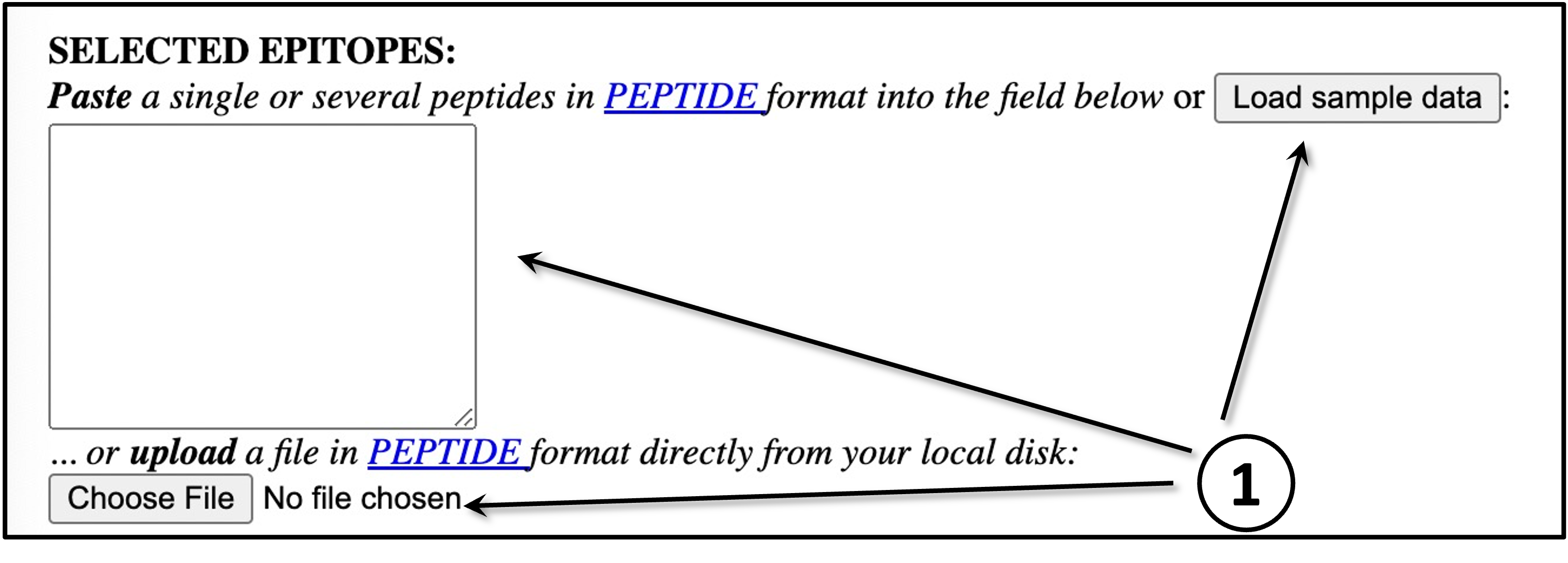
EPITOPES DATA
In this section, the user must define the input for the construction server following these
steps(Please note that the selected epitopes must match the selected alleles):
1)
Provide the input data by means of pasting the data into the blank field, uploading it
using the "Choose File" button or by loading sample data using the "Load Data" button. All the
input sequences must be in one-letter amino acid code. The alphabet is as follows (case
sensitive):
A C D E F G H I K L M N P Q R S T V W Y and X (unknown)
Any other symbol will let the processing stop. At most 10 selected epitopes are allowed per
submission because of the running time limitation; each sequence must be not more than 14 amino
acids long and not less than 8 amino acids long. PolyVaccine-1.0 will predict the peptide-MHC binder with 9 amino acids long.
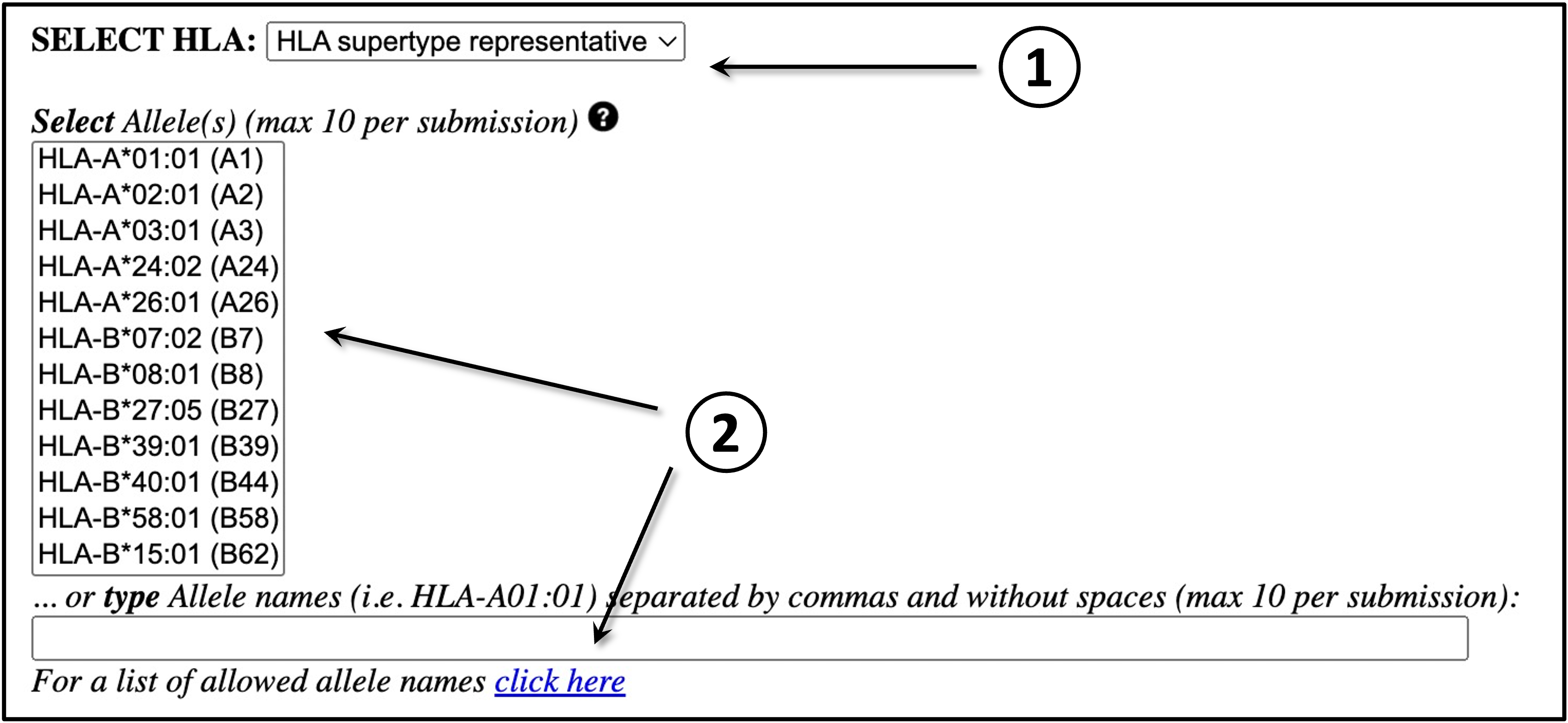
MHC SELECTION
Here, the user must define which MHC(s) molecule(s) the input data is going to be predicted
against:
1)
First, select the HLA/MHC supertype family.
2)
After selecting the MHC family, the user will be able to select a single or multiple MHC
molecules from the updated "Select Allele(s)" list. On the other hand, the user may opt to
directly type the MHC names in the provided blank field (separated by commas and without blank
spaces); if this is the case, there will be no need to select an MHC supertype familiy from the
drop-down menu. Click here for a list of MHC molecule
names (use the names in the first column). Please note that a maximum of 10 MHC types is allowed
per submission.
OPTIONAL LINKERS DATA
In this section, the user has chance to provide the preferred linkers for the vaccine
consturction:
1)
Provide the input data by means of pasting the data into the blank field. All the input
sequences must be in one-letter amino acid code. The alphabet is as follows (case
sensitive):
A C D E F G H I K L M N P Q R S T V W Y and X (unknown)
Any other symbol will let the processing stop. At most 10 selected linkers are allowed per
submission; each sequence must be not more than 9 amino acids long. The input data has to be one selected linker per line.
Only the big step Greedy search will be applied on with the preferred linkers. If the user want to keep optimizing the polytope vaccine, please click on the button 'Improve with the slow Monte Carlo method' on the first ouput page.
ADDITIONAL CONFIGURATION
In this section, the user may define additional parameters to further customize the run:
1)
Define the weight the user would penalize the cost function for the linkers that longer than the threshold.
2)
Define the threshold for the length of the linkers.

SUBMISSION
After the user has finished the "EPITOPES DATA", "MHC SELECTION" and "OPTIONAL LINKERS
DATA", "ADDITIONAL CONFIGURATION" steps, the submission can now be done. To do so, the user can click on "Submit" to submit
the job to the processing server, or click on "Clear fields" to clear the page and start over.
The status of your job (either 'queued' or 'running') will be displayed and constantly updated
until it terminates and the server output appears in the browser window.
After the server has finished running the corresponding predictions, an
output page will be delivered to the user. A description of the output format can be found at
output format
At any time during the wait you may enter your e-mail address and simply leave the window. Your
job will continue; when it terminates you will be notified by e-mail with a URL to your results.
They will be stored on the server for 24 hours.

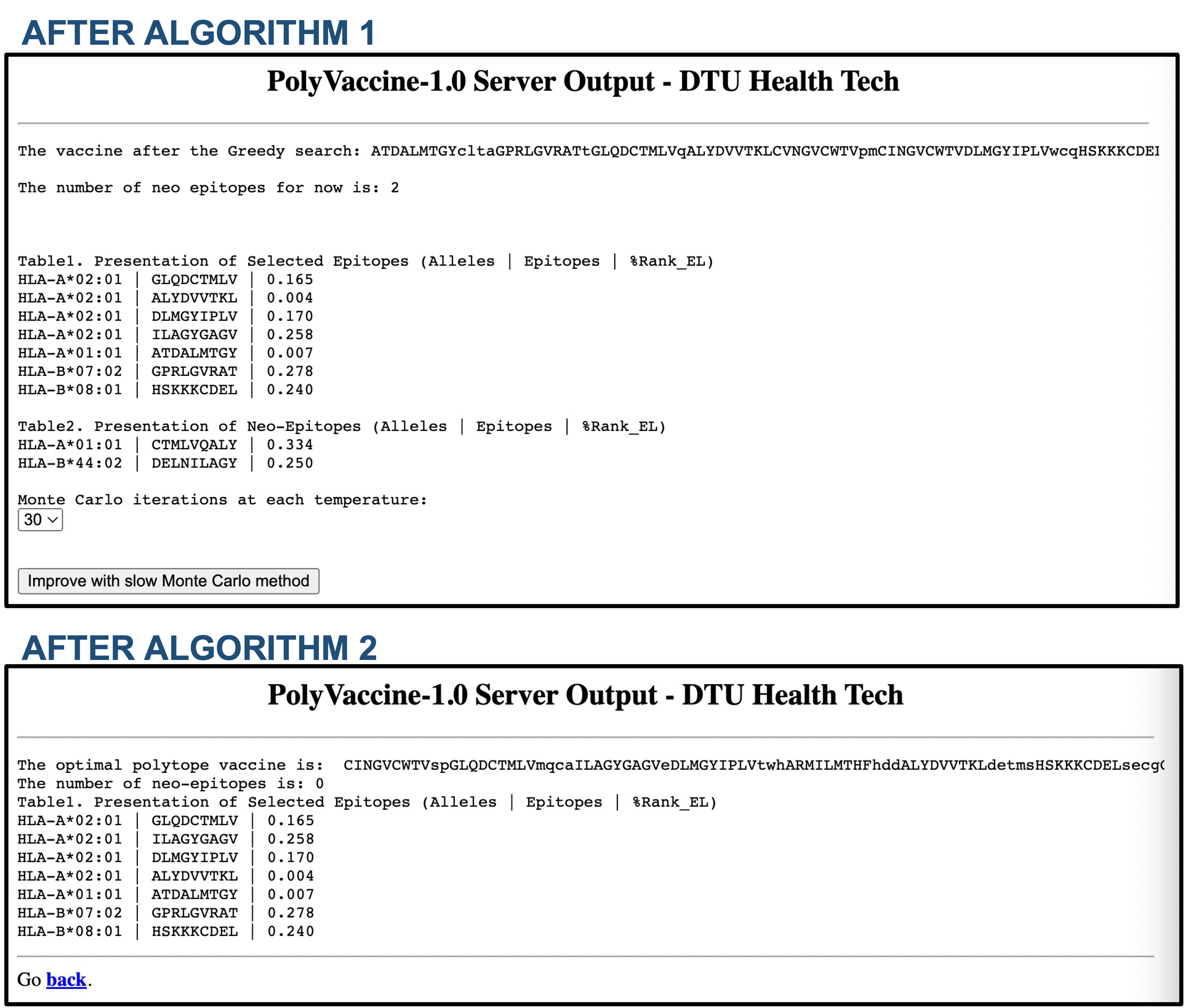
AFTER THE ALGORITHM 1
After the user has clicked on "Submit" to submit
the job to the processing server, the first output page will be given after a while. Either the vaccine with 0 neo-epitope will be produced or the vaccine with more than 0 neo-epitope will be shown so that the ALGORITHM 2(Monte Carlo) can be applied to keep optimizing the vaccine.
1)
The finite steps at each temperature in ALGORITHM 2 can be selected (30 or 40 or 50).
2)
Click on "Improve with slow Monte Carlo method" to keep optimizing the vaccine if you want. It's quite time comsuming, so be patient.
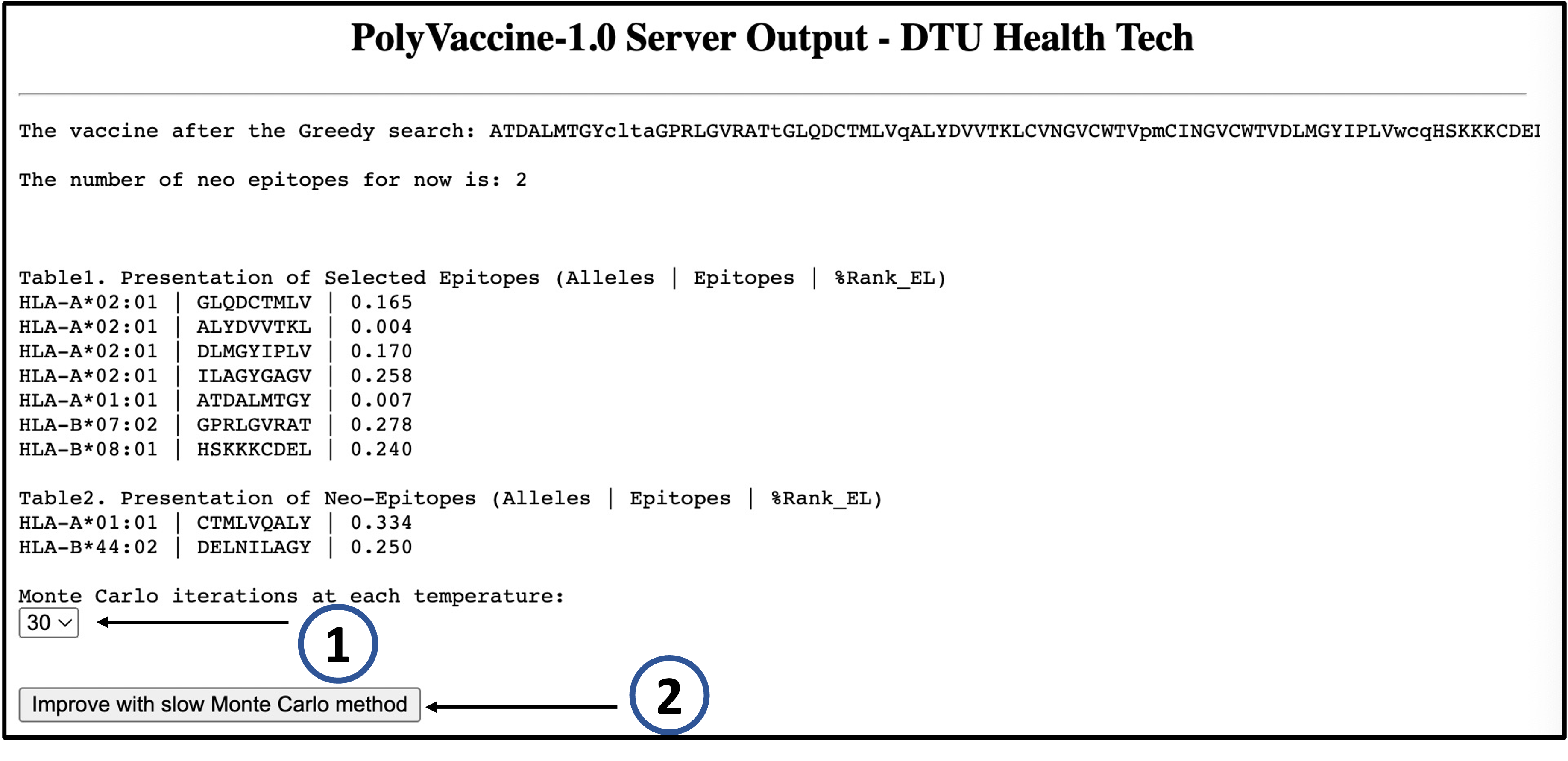
EXAMPLE
For the following EPITOPES(from HCV) input example:
CINGVCWTV
ALYDVVTKL
DLMGYIPLV
GLQDCTMLV
CVNGVCWTV
HSKKKCDEL
ATDALMTGY
GPRLGVRAT
ARMILMTHF
ILAGYGAGV
With parameters:
Allele: HLA-A02:01,HLA-A01:01,HLA-A03:01,HLA-A24:02,HLA-A11:01,HLA-B07:02,HLA-B51:01,HLA-B08:01,HLA-B35:01,HLA-B44:02
OPTINAL LINKERS: no preferred linkers were provided, so the program generated linkers automatically.
ADDITIONAL CONFIGURATION: with default value means no cost function for the linkers.
PolyVaccine-1.0 will return the following outputs (selected "30" and clicked "Improve with slow Monte Carlo method" after ALGORITHM 1):