After the table, the whole sequence is printed alongside a summary of the predicted glycation sites and their positions.
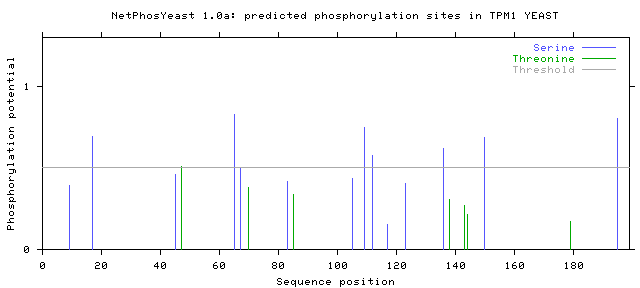
Finally, if the 'Generate graphics' button has been checked, the server displays a figure in GIF showing a plot of the score for each serine or threonine residue against the sequence position of that residue.
>TPM1_YEAST 199 amino acids
#
# netphosyeast-1.0a prediction results
#
# Sequence # x Context Score Kinase Answer
# -------------------------------------------------------------------
# TPM1_YEAST 9 S REKLSNLKL 0.390 main .
# TPM1_YEAST 17 S LEAESWQEK 0.692 main YES
# TPM1_YEAST 45 S NQIKSLTVK 0.460 main .
# TPM1_YEAST 47 T IKSLTVKNQ 0.508 main YES
# TPM1_YEAST 65 S EAGLSDSKQ 0.829 main YES
# TPM1_YEAST 67 S GLSDSKQTE 0.502 main YES
# TPM1_YEAST 70 T DSKQTEQDN 0.383 main .
# TPM1_YEAST 83 S NQIKSLTVK 0.416 main .
# TPM1_YEAST 85 T IKSLTVKNH 0.338 main .
# TPM1_YEAST 105 S ELAESKQLS 0.434 main .
# TPM1_YEAST 109 S SKQLSEDSH 0.750 main YES
# TPM1_YEAST 112 S LSEDSHHLQ 0.578 main YES
# TPM1_YEAST 117 S HHLQSNNDN 0.152 main .
# TPM1_YEAST 123 S NDNFSKKNQ 0.406 main .
# TPM1_YEAST 136 S DLEESDTKL 0.620 main YES
# TPM1_YEAST 138 T EESDTKLKE 0.308 main .
# TPM1_YEAST 143 T KLKETTEKL 0.269 main .
# TPM1_YEAST 144 T LKETTEKLR 0.213 main .
# TPM1_YEAST 150 S KLRESDLKA 0.685 main YES
# TPM1_YEAST 179 T NEELTVKYE 0.172 main .
# TPM1_YEAST 195 S EIAASLENL 0.803 main YES
#
MDKIREKLSNLKLEAESWQEKYEELKEKNKDLEQENVEKENQIKSLTVKN # 50
QQLEDEIEKLEAGLSDSKQTEQDNVEKENQIKSLTVKNHQLEEEIEKLEA # 100
ELAESKQLSEDSHHLQSNNDNFSKKNQQLEEDLEESDTKLKETTEKLRES # 150
DLKADQLERRVAALEEQREEWERKNEELTVKYEDAKKELDEIAASLENL # 200
%1 ................S.............................T... # 50
%1 ..............S.S................................. # 100
%1 ........S..S.......................S.............S # 150
%1 ............................................S....