DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
The NetPhosYeast 1.0 server predicts serine and threonine phosphorylation sites in yeast proteins. This service is closely related to NetPhos and NetPhosK.
Sequence submission: paste the sequence(s) and/or upload a local file
Restrictions:
At most 2,000 sequences and 200,000 amino acids per submission;
each sequence not longer than 6,000 amino acids.
Confidentiality:
The sequences are kept confidential and will be deleted
after processing.
For publication of results, please cite:
NetPhosYeast: Prediction of protein phosphorylation sites in yeast.
Christian Ravnsborg Ingrell, Martin Lee Miller, Ole Nørregaard
Jensen and Nikolaj Blom.
Accepted for publication in Bioinformatics, 2007.
All the other symbols will be converted to X before processing. The sequences can be input in the following two ways:
Both ways can be employed at the same time: all the specified sequences will
be processed. However, there may be not more than 2,000 sequences and
200,000 amino acids
in total in one submission. The sequences
longer than 6,000 amino acids are not allowed.
At any time during the wait you may enter your e-mail address and simply leave the window. Your job will continue; you will be notified by e-mail when it has terminated. The e-mail message will contain the URL under which the results are stored; they will remain on the server for 24 hours for you to collect them.
After the table, the whole sequence is printed alongside a summary of the predicted glycation sites and their positions.
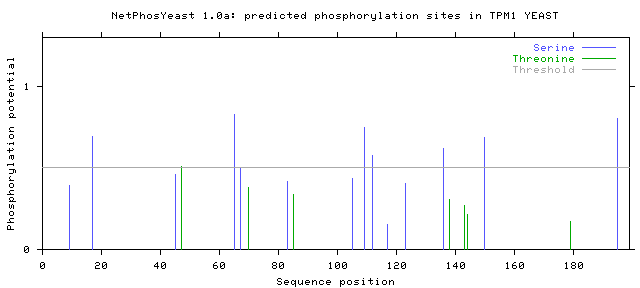
Finally, if the 'Generate graphics' button has been checked, the server displays a figure in GIF showing a plot of the score for each serine or threonine residue against the sequence position of that residue.
>TPM1_YEAST 199 amino acids
#
# netphosyeast-1.0a prediction results
#
# Sequence # x Context Score Kinase Answer
# -------------------------------------------------------------------
# TPM1_YEAST 9 S REKLSNLKL 0.390 main .
# TPM1_YEAST 17 S LEAESWQEK 0.692 main YES
# TPM1_YEAST 45 S NQIKSLTVK 0.460 main .
# TPM1_YEAST 47 T IKSLTVKNQ 0.508 main YES
# TPM1_YEAST 65 S EAGLSDSKQ 0.829 main YES
# TPM1_YEAST 67 S GLSDSKQTE 0.502 main YES
# TPM1_YEAST 70 T DSKQTEQDN 0.383 main .
# TPM1_YEAST 83 S NQIKSLTVK 0.416 main .
# TPM1_YEAST 85 T IKSLTVKNH 0.338 main .
# TPM1_YEAST 105 S ELAESKQLS 0.434 main .
# TPM1_YEAST 109 S SKQLSEDSH 0.750 main YES
# TPM1_YEAST 112 S LSEDSHHLQ 0.578 main YES
# TPM1_YEAST 117 S HHLQSNNDN 0.152 main .
# TPM1_YEAST 123 S NDNFSKKNQ 0.406 main .
# TPM1_YEAST 136 S DLEESDTKL 0.620 main YES
# TPM1_YEAST 138 T EESDTKLKE 0.308 main .
# TPM1_YEAST 143 T KLKETTEKL 0.269 main .
# TPM1_YEAST 144 T LKETTEKLR 0.213 main .
# TPM1_YEAST 150 S KLRESDLKA 0.685 main YES
# TPM1_YEAST 179 T NEELTVKYE 0.172 main .
# TPM1_YEAST 195 S EIAASLENL 0.803 main YES
#
MDKIREKLSNLKLEAESWQEKYEELKEKNKDLEQENVEKENQIKSLTVKN # 50
QQLEDEIEKLEAGLSDSKQTEQDNVEKENQIKSLTVKNHQLEEEIEKLEA # 100
ELAESKQLSEDSHHLQSNNDNFSKKNQQLEEDLEESDTKLKETTEKLRES # 150
DLKADQLERRVAALEEQREEWERKNEELTVKYEDAKKELDEIAASLENL # 200
%1 ................S.............................T... # 50
%1 ..............S.S................................. # 100
%1 ........S..S.......................S.............S # 150
%1 ............................................S....
1University of Southern Denmark,
Campusvej 55, DK-5230, Odense M, Denmark
2Center for Biological Sequence Analysis, BioCentrum-DTU,
The Technical University of Denmark, DK-2800 Lyngby, Denmark
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: