Output format
Classical format
For each input sequence the following is shown (see the example below):
- FASTA-like header line: a line showing the sequence name and length.
- Prediction lines: one line per residue and kinase, with six columns
in the form:
- Sequence - the sequence name;
- # - the position of the residue in the
sequence;
- x - the residue in one-letter code;
- Context - the sequence context of the
residue, shown as a 9-residue subsequence centered on the residue;
- Score - the prediction score (a value in the range [0.000-1.000];
the scores above 0.500 indicate positive predictions);
- Kinase - the active kinase or the string "unsp" for non-specific
prediction (as in NetPhos 2.0);
- Answer - the string "YES" for positive predictions,
else a dot.
- Sequence - the input sequence as processed by NetPhos, with an
overview of the positions of the predicted sites.
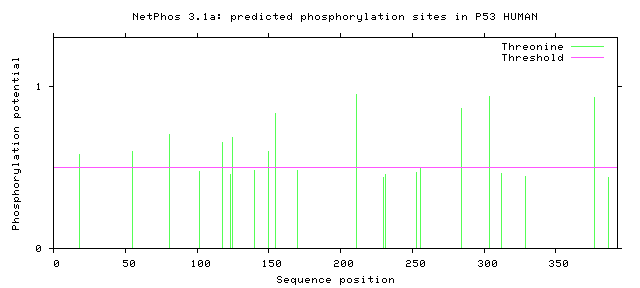
- Graphics - a plot of scores illustrating the predictions. NOTE: for
each residue only the highest score is shown.
GFF
The output in GFF (
GFF version 2)
provides essentially the same information as the classical format described
above. The only differences, apart from the syntax, are as follows:
- the sequence context of the residues is not provided
- the positive predictions are indicated by "Y" (not "YES")
This option has been provided for the benefit of the users who have access to
software parsing GFF.
Example
The NetPhos 3.1 output for the UniProtKB entry P53_HUMAN
(P04637).
The predictions are for tyrosine only, showing only the highest scoring
prediction for each residue and skipping all the predictions with scores
lower than 0.250.
Classical format
>P53_HUMAN 393 amino acids
#
# netphos-3.1b prediction results
#
# Sequence # x Context Score Kinase Answer
# -------------------------------------------------------------------
# P53_HUMAN 18 T LSQETFSDL 0.582 CKI YES
# P53_HUMAN 55 T EQWFTEDPG 0.598 CKII YES
# P53_HUMAN 81 T PAAPTPAAP 0.704 unsp YES
# P53_HUMAN 102 T PSQKTYQGS 0.472 cdc2 .
# P53_HUMAN 118 T LHSGTAKSV 0.654 PKC YES
# P53_HUMAN 123 T AKSVTCTYS 0.455 GSK3 .
# P53_HUMAN 125 T SVTCTYSPA 0.684 PKC YES
# P53_HUMAN 140 T QLAKTCPVQ 0.479 cdc2 .
# P53_HUMAN 150 T WVDSTPPPG 0.599 unsp YES
# P53_HUMAN 155 T PPPGTRVRA 0.829 unsp YES
# P53_HUMAN 170 T SQHMTEVVR 0.482 unsp .
# P53_HUMAN 211 T DDRNTFRHS 0.951 unsp YES
# P53_HUMAN 230 T GSDCTTIHY 0.438 GSK3 .
# P53_HUMAN 231 T SDCTTIHYN 0.454 GSK3 .
# P53_HUMAN 253 T RPILTIITL 0.467 CaM-II .
# P53_HUMAN 256 T LTIITLEDS 0.492 cdc2 .
# P53_HUMAN 284 T RDRRTEEEN 0.865 unsp YES
# P53_HUMAN 304 T PPGSTKRAL 0.939 unsp YES
# P53_HUMAN 312 T LPNNTSSSP 0.459 CaM-II .
# P53_HUMAN 329 T GEYFTLQIR 0.443 GSK3 .
# P53_HUMAN 377 T KGQSTSRHK 0.932 unsp YES
# P53_HUMAN 387 T LMFKTEGPD 0.440 GSK3 .
#
MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDI # 50
EQWFTEDPGPDEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQ # 100
KTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKTCPVQLWVDST # 150
PPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGN # 200
LRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRP # 250
ILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP # 300
PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALEL # 350
KDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD # 400
%1 .................T................................ # 50
%1 ....T.........................T................... # 100
%1 .................T......T........................T # 150
%1 ....T............................................. # 200
%1 ..........T....................................... # 250
%1 .................................T................ # 300
%1 ...T.............................................. # 350
%1 ..........................T................
GFF
The corresponding output in GFF (the graph is not shown again):
##gff-version 2
##source-version netphos-3.1b
##date 2016-07-12
##Type Protein P53_HUMAN
##Protein P53_HUMAN
##MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGP
##DEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAK
##SVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHE
##RCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNS
##SCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP
##PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPG
##GSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD
##end-Protein
# seqname source feature start end score N/A ?
# ---------------------------------------------------------------------------
P53_HUMAN netphos-3.1b phos-CKI 18 18 0.582 . . YES
P53_HUMAN netphos-3.1b phos-CKII 55 55 0.598 . . YES
P53_HUMAN netphos-3.1b phos-unsp 81 81 0.704 . . YES
P53_HUMAN netphos-3.1b phos-cdc2 102 102 0.472 . . .
P53_HUMAN netphos-3.1b phos-PKC 118 118 0.654 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 123 123 0.455 . . .
P53_HUMAN netphos-3.1b phos-PKC 125 125 0.684 . . YES
P53_HUMAN netphos-3.1b phos-cdc2 140 140 0.479 . . .
P53_HUMAN netphos-3.1b phos-unsp 150 150 0.599 . . YES
P53_HUMAN netphos-3.1b phos-unsp 155 155 0.829 . . YES
P53_HUMAN netphos-3.1b phos-unsp 170 170 0.482 . . .
P53_HUMAN netphos-3.1b phos-unsp 211 211 0.951 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 230 230 0.438 . . .
P53_HUMAN netphos-3.1b phos-GSK3 231 231 0.454 . . .
P53_HUMAN netphos-3.1b phos-CaM-II 253 253 0.467 . . .
P53_HUMAN netphos-3.1b phos-cdc2 256 256 0.492 . . .
P53_HUMAN netphos-3.1b phos-unsp 284 284 0.865 . . YES
P53_HUMAN netphos-3.1b phos-unsp 304 304 0.939 . . YES
P53_HUMAN netphos-3.1b phos-CaM-II 312 312 0.459 . . .
P53_HUMAN netphos-3.1b phos-GSK3 329 329 0.443 . . .
P53_HUMAN netphos-3.1b phos-unsp 377 377 0.932 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 387 387 0.440 . . .
