DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
 |
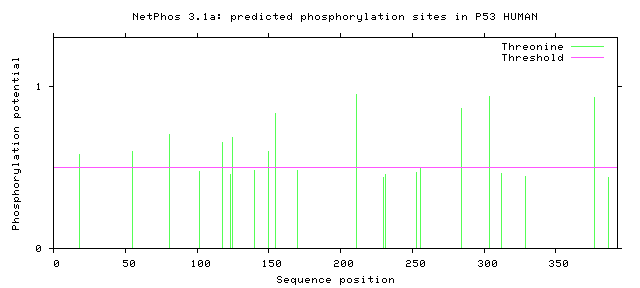
ATM, CKI, CKII, CaM-II, DNAPK, EGFR, GSK3, INSR, PKA, PKB, PKC, PKG, RSK, SRC, cdc2, cdk5 and p38MAPK.
Sequence submission: paste the sequence(s) and/or upload a local file
Restrictions:
At most 2000 sequences and 200,000 amino acids
per submission; each sequence not less than 15 and not more than 4,000 amino
acids.
Confidentiality:
The sequences are kept confidential and will be
deleted after processing.
For publication of results, please cite:
Generic predictions:
Sequence- and structure-based prediction of eukaryotic protein
phosphorylation sites.
Blom, N., Gammeltoft, S., and Brunak, S.
Journal of Molecular Biology: 294(5): 1351-1362, 1999.
PMID: 10600390
Kinase specific predictions:
Prediction of post-translational glycosylation and phosphorylation of
proteins from the amino acid sequence.
Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S.
Proteomics: Jun;4(6):1633-49, review 2004.
PMID: 15174133
The characters not in the one-letter amino acid code e.g. letters like 'B' or 'Z', digits and other non-alphabetic symbols will all be converted to X before processing; they will be accepted but not used when making the predictions. White space within the sequences, if any, will be ignored. The sequences can be input in the following two ways:
At any time during the wait you may enter your e-mail address and simply
leave the window. Your job will continue; you will be notified by e-mail
when it has terminated. The e-mail message will contain the URL under which
the results are stored; they will remain on the server for 24 hours for you
to collect them.
>P53_HUMAN 393 amino acids
#
# netphos-3.1b prediction results
#
# Sequence # x Context Score Kinase Answer
# -------------------------------------------------------------------
# P53_HUMAN 18 T LSQETFSDL 0.582 CKI YES
# P53_HUMAN 55 T EQWFTEDPG 0.598 CKII YES
# P53_HUMAN 81 T PAAPTPAAP 0.704 unsp YES
# P53_HUMAN 102 T PSQKTYQGS 0.472 cdc2 .
# P53_HUMAN 118 T LHSGTAKSV 0.654 PKC YES
# P53_HUMAN 123 T AKSVTCTYS 0.455 GSK3 .
# P53_HUMAN 125 T SVTCTYSPA 0.684 PKC YES
# P53_HUMAN 140 T QLAKTCPVQ 0.479 cdc2 .
# P53_HUMAN 150 T WVDSTPPPG 0.599 unsp YES
# P53_HUMAN 155 T PPPGTRVRA 0.829 unsp YES
# P53_HUMAN 170 T SQHMTEVVR 0.482 unsp .
# P53_HUMAN 211 T DDRNTFRHS 0.951 unsp YES
# P53_HUMAN 230 T GSDCTTIHY 0.438 GSK3 .
# P53_HUMAN 231 T SDCTTIHYN 0.454 GSK3 .
# P53_HUMAN 253 T RPILTIITL 0.467 CaM-II .
# P53_HUMAN 256 T LTIITLEDS 0.492 cdc2 .
# P53_HUMAN 284 T RDRRTEEEN 0.865 unsp YES
# P53_HUMAN 304 T PPGSTKRAL 0.939 unsp YES
# P53_HUMAN 312 T LPNNTSSSP 0.459 CaM-II .
# P53_HUMAN 329 T GEYFTLQIR 0.443 GSK3 .
# P53_HUMAN 377 T KGQSTSRHK 0.932 unsp YES
# P53_HUMAN 387 T LMFKTEGPD 0.440 GSK3 .
#
MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDI # 50
EQWFTEDPGPDEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQ # 100
KTYQGSYGFRLGFLHSGTAKSVTCTYSPALNKMFCQLAKTCPVQLWVDST # 150
PPPGTRVRAMAIYKQSQHMTEVVRRCPHHERCSDSDGLAPPQHLIRVEGN # 200
LRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNSSCMGGMNRRP # 250
ILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP # 300
PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALEL # 350
KDAQAGKEPGGSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD # 400
%1 .................T................................ # 50
%1 ....T.........................T................... # 100
%1 .................T......T........................T # 150
%1 ....T............................................. # 200
%1 ..........T....................................... # 250
%1 .................................T................ # 300
%1 ...T.............................................. # 350
%1 ..........................T................
##gff-version 2
##source-version netphos-3.1b
##date 2016-07-12
##Type Protein P53_HUMAN
##Protein P53_HUMAN
##MEEPQSDPSVEPPLSQETFSDLWKLLPENNVLSPLPSQAMDDLMLSPDDIEQWFTEDPGP
##DEAPRMPEAAPPVAPAPAAPTPAAPAPAPSWPLSSSVPSQKTYQGSYGFRLGFLHSGTAK
##SVTCTYSPALNKMFCQLAKTCPVQLWVDSTPPPGTRVRAMAIYKQSQHMTEVVRRCPHHE
##RCSDSDGLAPPQHLIRVEGNLRVEYLDDRNTFRHSVVVPYEPPEVGSDCTTIHYNYMCNS
##SCMGGMNRRPILTIITLEDSSGNLLGRNSFEVRVCACPGRDRRTEEENLRKKGEPHHELP
##PGSTKRALPNNTSSSPQPKKKPLDGEYFTLQIRGRERFEMFRELNEALELKDAQAGKEPG
##GSRAHSSHLKSKKGQSTSRHKKLMFKTEGPDSD
##end-Protein
# seqname source feature start end score N/A ?
# ---------------------------------------------------------------------------
P53_HUMAN netphos-3.1b phos-CKI 18 18 0.582 . . YES
P53_HUMAN netphos-3.1b phos-CKII 55 55 0.598 . . YES
P53_HUMAN netphos-3.1b phos-unsp 81 81 0.704 . . YES
P53_HUMAN netphos-3.1b phos-cdc2 102 102 0.472 . . .
P53_HUMAN netphos-3.1b phos-PKC 118 118 0.654 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 123 123 0.455 . . .
P53_HUMAN netphos-3.1b phos-PKC 125 125 0.684 . . YES
P53_HUMAN netphos-3.1b phos-cdc2 140 140 0.479 . . .
P53_HUMAN netphos-3.1b phos-unsp 150 150 0.599 . . YES
P53_HUMAN netphos-3.1b phos-unsp 155 155 0.829 . . YES
P53_HUMAN netphos-3.1b phos-unsp 170 170 0.482 . . .
P53_HUMAN netphos-3.1b phos-unsp 211 211 0.951 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 230 230 0.438 . . .
P53_HUMAN netphos-3.1b phos-GSK3 231 231 0.454 . . .
P53_HUMAN netphos-3.1b phos-CaM-II 253 253 0.467 . . .
P53_HUMAN netphos-3.1b phos-cdc2 256 256 0.492 . . .
P53_HUMAN netphos-3.1b phos-unsp 284 284 0.865 . . YES
P53_HUMAN netphos-3.1b phos-unsp 304 304 0.939 . . YES
P53_HUMAN netphos-3.1b phos-CaM-II 312 312 0.459 . . .
P53_HUMAN netphos-3.1b phos-GSK3 329 329 0.443 . . .
P53_HUMAN netphos-3.1b phos-unsp 377 377 0.932 . . YES
P53_HUMAN netphos-3.1b phos-GSK3 387 387 0.440 . . .
Phospho.ELM is becoming the primary resource for phosphorylation data. CBS continues to contribute to the database alongside many other research groups.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: