Graphics in PostScript
# Predictions for N-Glycosylation sites in 1 sequence Name: CBG_HUMAN Length: 405
(Sequence) Asn-Xaa-Ser/Thr sequons (including Asn-Pro-Ser/Thr) are shown in blue. Asparagines predicted to be N-glycosylated are shown in red. Note that not all sequons are predicted glycosylated.
MPLLLYTCLLWLPTSGLWTVQAMDPNAAYVNMSNHHRGLASANVDFAFSLYKHLVALSPKKNIFISPVSISMALAMLSLG 80 TCGHTRAQLLQGLGFNLTERSETEIHQGFQHLHQLFAKSDTSLEMTMGNALFLDGSLELLESFSADIKHYYESEVLAMNF 160 QDWATASRQINSYVKNKTQGKIVDLFSGLDSPAILVLVNYIFFKGTWTQPFDLASTREENFYVDETTVVKVPMMLQSSTI 240 SYLHDSELPCQLVQMNYVGNGTVFFILPDKGKMNTVIAALSRDTINRWSAGLTSSQVDLYIPKVTISGVYDLGDVLEEMG 320 IADLFTNQANFSRITQDAQLKSSKVVHKAVLQLNEEGVDTAGSTGVTLNLTSKPIILRFNQPFIIMIFDHFTWSSLFLAR 400 VMNPV (Annotation line) `N' represents a predicted N-glycosylation site. `n' represents an Asn with a positive score, but not occuring within an Asn-Xaa-Ser/Thr sequon
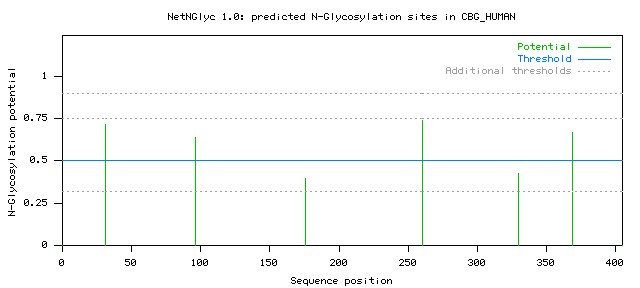
..............................N................................................. 80 ...............N................................................................ 160 ................................................................................ 240 ...................N............................................................ 320 ................................................N............................... 400 ..... (Threshold=0.5) -------------------------------------------------------------------------------- SeqName Position Potential Jury NGlyc agreement result -------------------------------------------------------------------------------- CBG_HUMAN 31 NMSN 0.7166 (9/9) ++ <-- Predicted as N-glycosylated (++) CBG_HUMAN 96 NLTE 0.6356 (8/9) + <-- Predicted as N-glycosylated (+) CBG_HUMAN 176 NKTQ 0.3941 (7/9) - <-- A negative site CBG_HUMAN 260 NGTV 0.7400 (9/9) ++ CBG_HUMAN 330 NFSR 0.4223 (7/9) - see below for CBG_HUMAN 369 NLTS 0.6684 (9/9) ++ more information --------------------------------------------------------------------------------
The graph illustrates predicted N-glyc sites across the protein chain (x-axis represents protein length from N- to C-terminal). A position with a potential (vertical lines) crossing the threshold (horizontal line at 0.5) is predicted glycosylated. Additional thresholds are shown at 0.32, 0.75 and 0.90 by horizontal dotted lines. Explained below. An Encapsulated postscript format of the graph is available for including in publications.
The Asn-Xaa-Ser/Thr sequon
Asn-Pro-Ser/Thr
Thresholds and confidence
+ Potential < 0.5 ++ Potential < 0.5 AND Jury agreement (9/9) OR Potential<0.75 +++ Potential < 0.75 AND Jury agreement ++++ Potential < 0.90 AND Jury agreementand non-glycosylated sites:
- Potential < 0.5 -- Potential < 0.5 AND Jury agreement (all nine > 0.5) --- Potential < 0.32 AND Jury agreement
Warnings and notes in the right margin
SEQUON ASN-XAA-SER/THR.If you request a prediction on all Asparagines (instead of the default to predict only on Asn-Xaa-Ser/Thr sequons), then this note will appear for Asparagine positions which do occur within the Asn-Xaa-Ser/Thr sequon.
WARNING: PRO-X1.Proline occurs just after the Asparagine residue. This makes it highly unlikely that the Asparagine is glycosylated, presumably due to conformational constraints.
WARNING: PRO-X2.Proline occurs at the 3rd position C-terminal to the Asparagine in question (2nd 'X' in NX[ST]X). This makes it somewhat unlikely that the Asparagine is glycosylated, but this condition is not as harsh as the PRO-X1 condition.