INPUT DATA
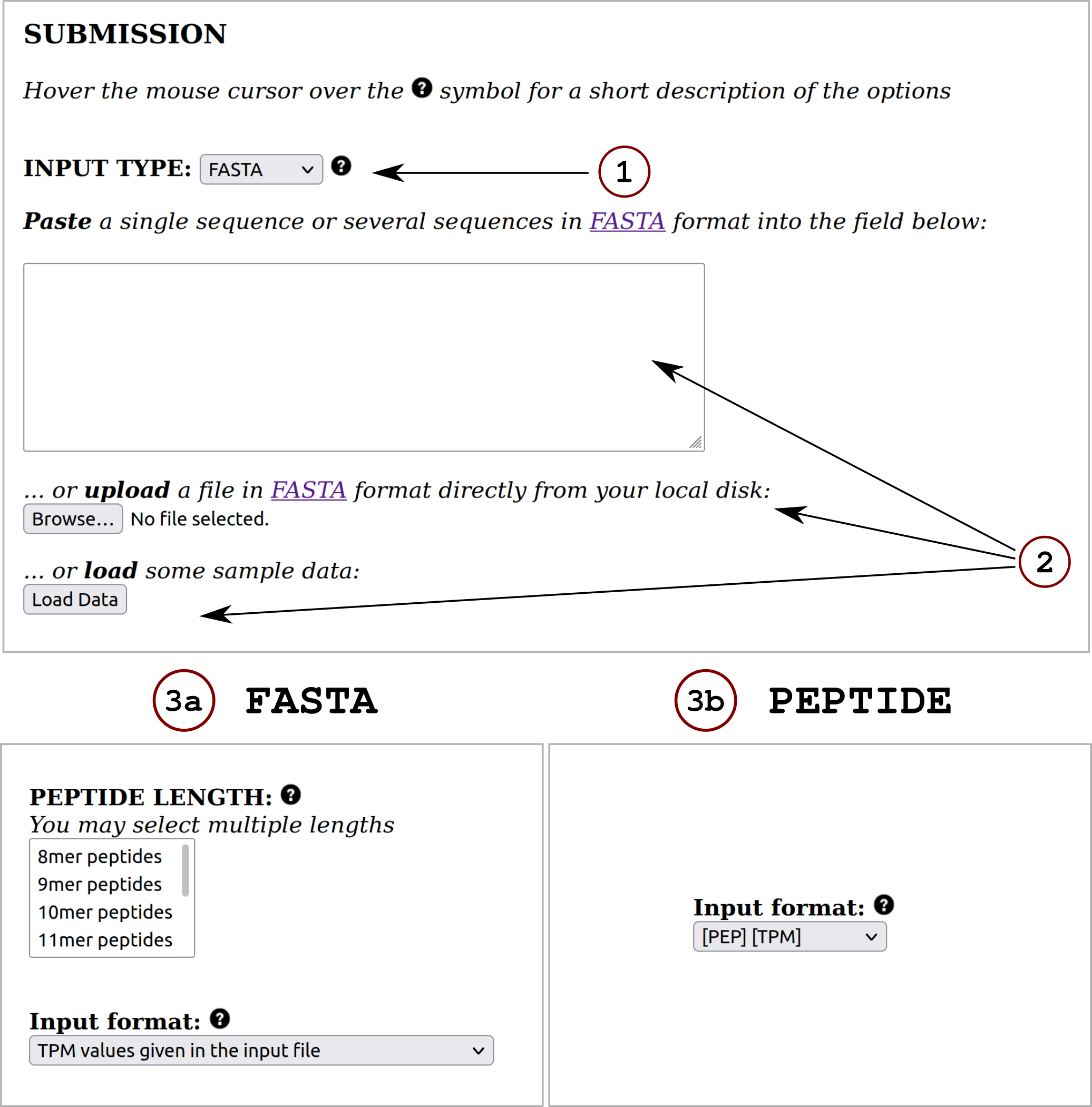
In this section, the user must define the input for the prediction server following these steps:1) Specify the desired input data type (FASTA or PEPTIDE) using the drop down menu.
2) Provide the input data by means of pasting the data into the blank field, uploading it using the "Choose File" button or by loading sample data using the "Load Data" button. All the input sequences must be in one-letter amino acid code. The alphabet is as follows (case sensitive):
A C D E F G H I K L M N P Q R S T V W Y and X (unknown)
Any other symbol will be converted to X before processing. At most 5000 sequences are allowed per submission; each sequence must be not more than 20,000 amino acids long and not less than 8 amino acids long.
3) Specify the input format (and peptide length(s) if required):
Sample input data is given after each of the listed options.
a. If FASTA was selected as input type, the user must choose the peptide length(s) the prediction server is going to work with. The input FASTA sequence is digested in overlapping peptides of the provided length(s) and the program predicts binding against all of them. By default input proteins are digested into 9-mer peptides.
Also, the user must decide between these 3 input format options:
- Lookup TPM values per peptide
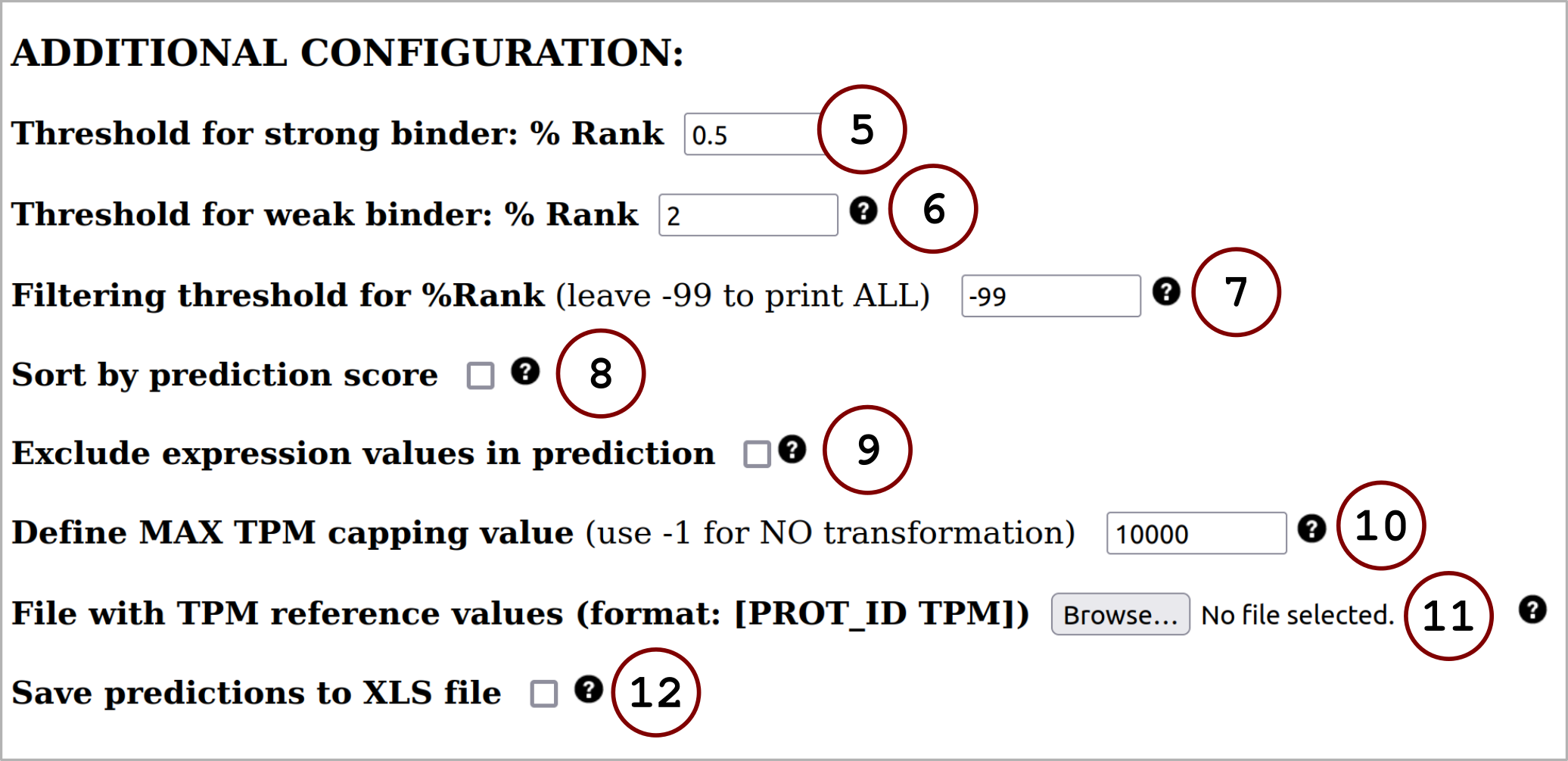
The digested peptides are searched against the HPA database. Gene expression value annotation for a given peptide is defined as the sum of the TPM values of all protein-coding transcripts containing the exact queried peptide. If a peptide is not found, the program will assign TPM=0.
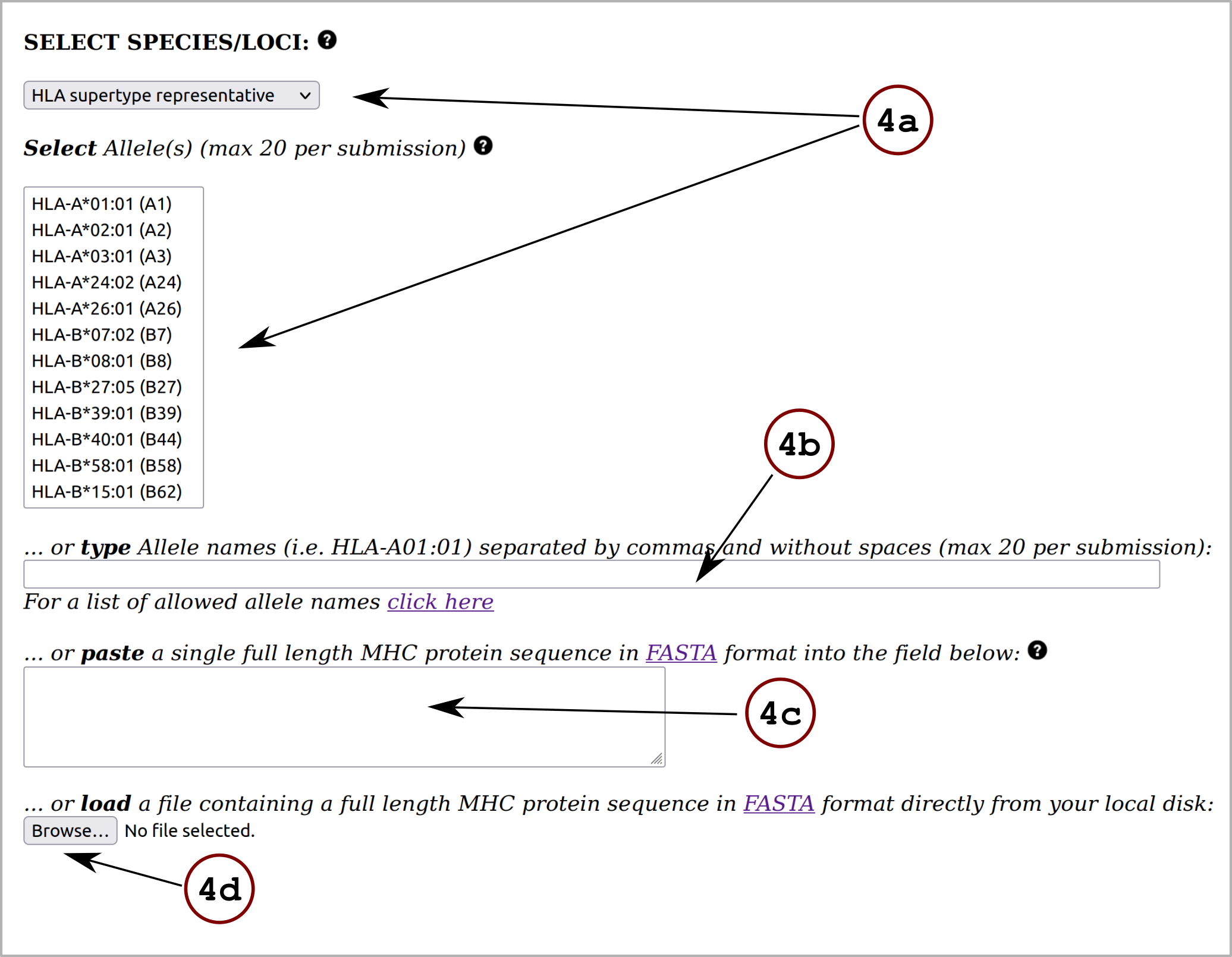
- Lookup TPM values per protein (using PROT_ID)
The PROT_ID is searched against the HPA database (or the user-specified expression database). Gene expression value annotation is the same for all digested peptides and is equal to the TPM value associated with the queried PROT_ID. If the PROT_ID is not found, the program will assign TPM=0.
Sample input data for options 1. & 2. :
>ENSP00000253039.4
MAGGEAGVTLGQPHLSRQDLTTLDVTKLTPLSHEVISRQATI...
- TPM values given in the input file
The digested peptides are assigned the TPM value annotated in the input file. This number should be a positive rational number. If this is not satisfied, the program will assign TPM=0.
>ENSP00000253039.4 TPM=80.3919983
MAGGEAGVTLGQPHLSRQDLTTLDVTKLTPLSHEVISRQATI...
b. If PEPTIDE was selected as input type, the user must select one of the following 3 input format options:
In all cases, the input format can contain an additional 2nd column that has numerical values. This column can be used to store scores (or target values) that can be futher on employed to compute evaluation metrics.
- [PEP] ([Score])
Peptides are searched against the HPA database. Gene expression value annotation works the same way as described above in a.1.
AAADIVNFL
AAADSIKIW
AAAHFYFEL
AAAPQLLIV
...
- [PEP] ([Score]) [PROT_ID]
PROT_ID(s) are searched against the HPA database (or the user-specified expression database). Gene expression value annotation is performed by assigning to each peptide its corresponding PROT_ID's TPM value. If the PROT_ID is not found, the program will assign TPM=0. For a list of the Ensembl protein IDs present in the HPA database (genome assembly GRCh38.p12, GENCODE v.28), click HERE.
AAADIVNFL ENSP00000404403.1
AAADSIKIW ENSP00000308179.4
AAAHFYFEL ENSP00000415612.1
AAAPQLLIV ENSP00000252593.6
...
- [PEP] ([Score]) [TPM]
The input is already in the required format for prediction.
AAADIVNFL 0.497551
AAADSIKIW 3.67981
AAAHFYFEL 8.54361
AAAPQLLIV 108.95
...
NOTE: If PEPTIDE was chosen as input type, peptide length selection is unnecessary and thus the corresponding selection box will directly not appear in the interface.