DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
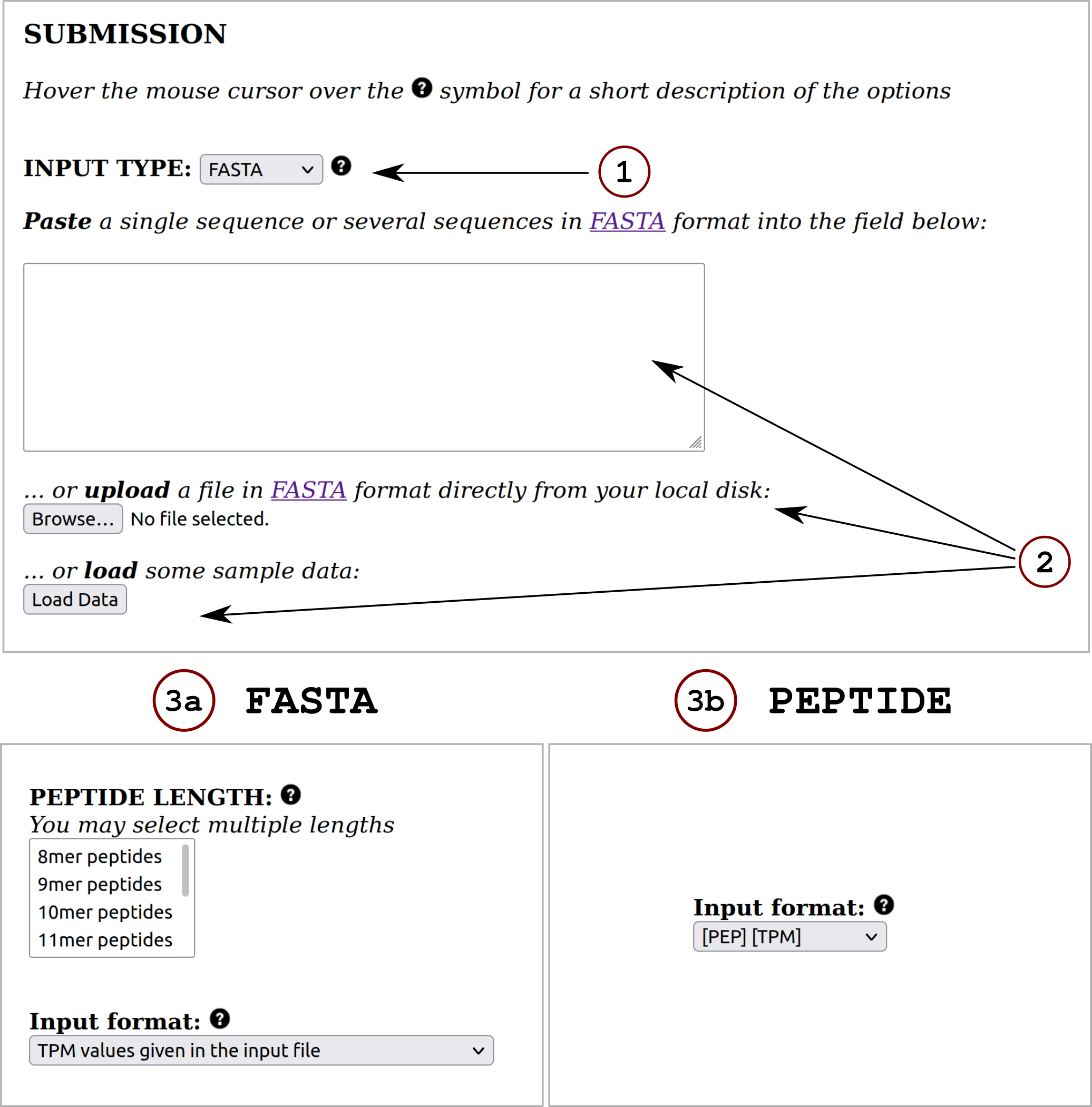
The NetMHCpanExp-1.0 server predicts binding of peptides to any human MHC-I molecule of known sequence using artificial neural networks (ANNs). This NetMHCpan-like method incorporates a new feature in the prediction of peptide-MHC class I binding: gene expression.
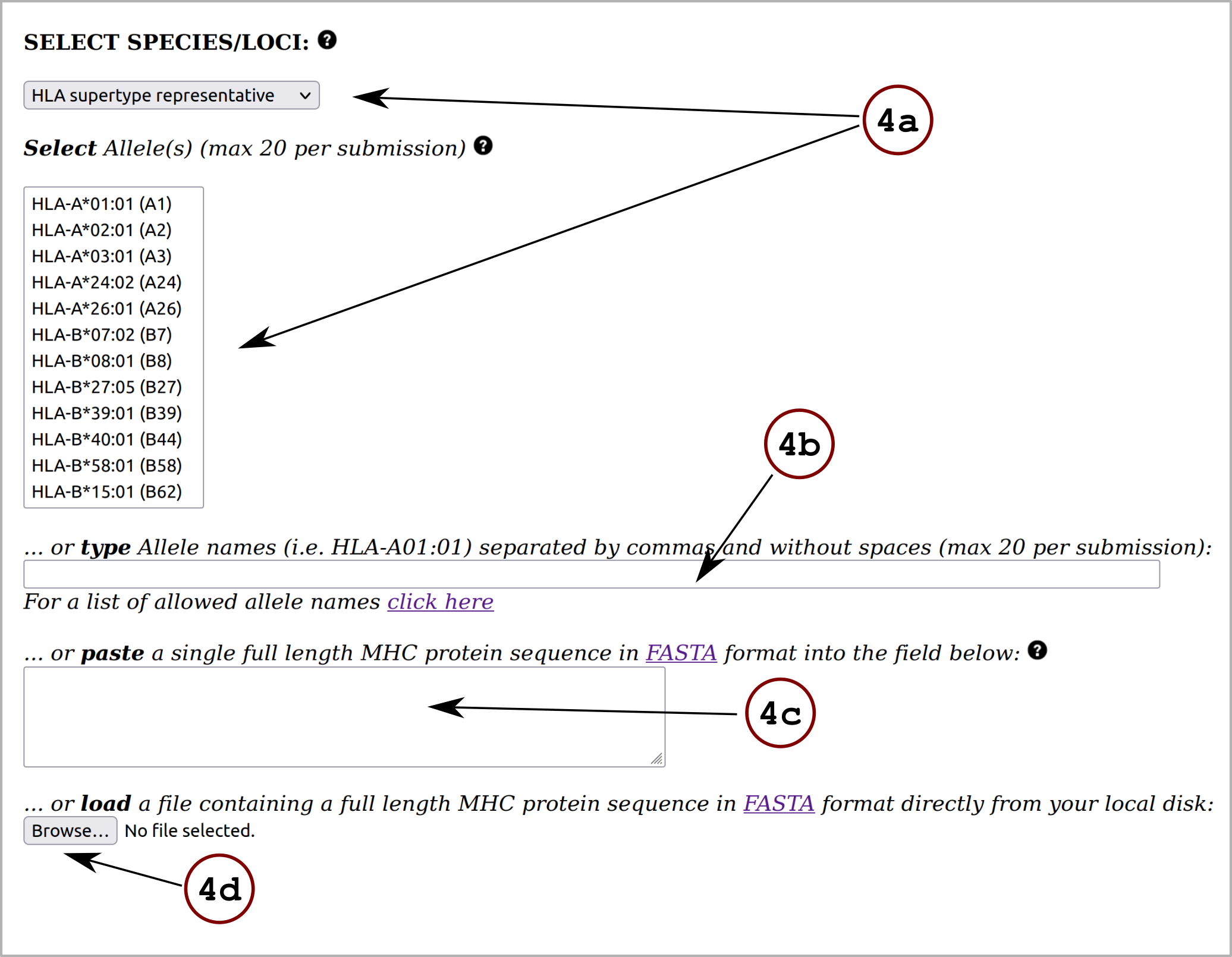
The method is trained on a combination of more than 670,000 quantitative Binding Affinity (BA) and Mass-Spectrometry Eluted Ligands (MS EL) peptides. The BA data covers 112 human MHC-I molecules (HLA-A, B, C and E) while the EL data covers 163 human MHC-I molecules (HLA-A, B, C and G). The user can obtain predictions to any custom human MHC class I molecule by uploading a full length MHC protein sequence. Predictions can be made for peptides of length 8 to 14.
This method, as well as its closest predecessor NetMHCpan-4.1, is built upon the algorithm NNAlign_MA which allows for simultaneous training and pseudo-labelling (automatic annotation) of multi-allelic data (MA), derived from MS experiments performed on cell lines or tissue samples. Additionally, in this case, the architecture of the algorithm NNAlign_MA was modified to accept gene expression values in the peptide encoding.
The current method was trained with MHC-I ligands derived from samples that were also assayed in RNA-Seq experiments. Finally, the training set was enlarged including MS EL ligands that originally lacked gene expression values by use of transcript abundance values derived from RNA-Seq experiments performed on 281 human tissue and blood cell samples deposited in the Human Protein Atlas (v. 20.0) database.
The user can make predictions for peptide (or protein) sequences with already annotated gene expression values (in Transcripts Per Millon). If the gene expression values are absent from the input data, the tool allows for automatic annotation of these values employing the Human Protein Atlas (v. 20.0) reference database. For more details, click on Instructions. Also, the user is enabled to make predictions without including the novel feature.
The server returns the likelihood of a peptide being a natural ligand of the selected MHC(s).
The project is a collaboration between the Bioinformatics Section at DTU HealthTech (see all available services) and LIAI.
For publication of results, please cite:
Data resources used to develop this server was obtained from:
Would you prefer to run NetMHCpanExp at your own site? NetMHCpanExp v. 1.0 is available as a stand-alone software package, with the same functionality as the service above. Ready-to-ship packages exist for the most common UNIX platforms. There is a download tap for academic users; other users are requested to contact DTU HealthTech Software Package Manager at health-software@dtu.dk.
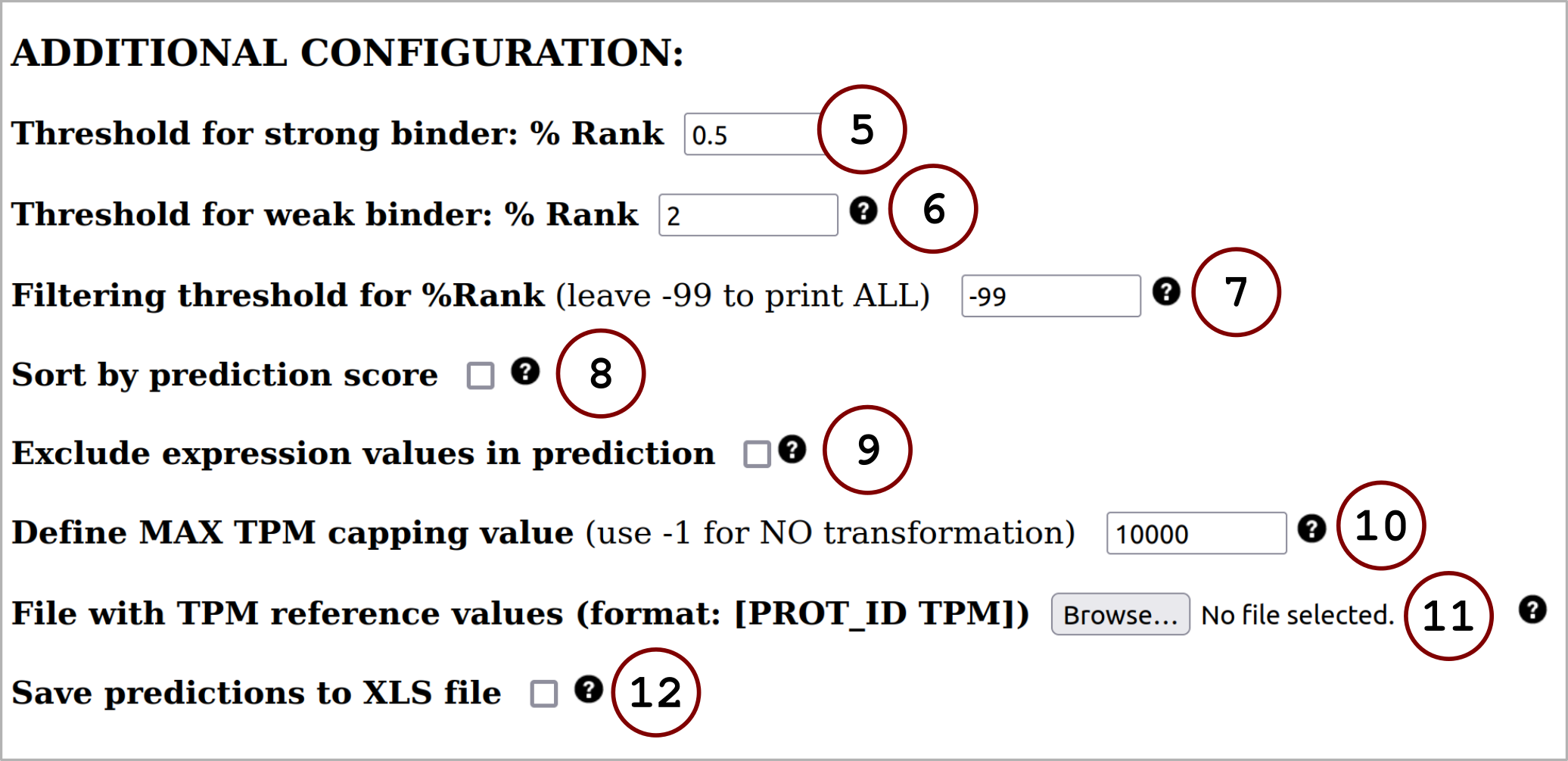

# NetMHCpanExp version 1.0 # Tmpdir made /var/www/webface/tmp/server/netmhcpanexp/6237B40000005ED4FDF6B89C/netMHCpanv9J7VL # Input is in FSA format # Peptide length 8,9,10,11 # Make predictions including expression HLA-A02:01 : Distance to training data 0.000 (using nearest neighbor HLA-A02:01) # Rank Threshold for Strong binding peptides 0.500 # Rank Threshold for Weak binding peptides 2.000 ------------------------------------------------------------------------------------------------------------ Pos HLA Peptide Core Of Gp Gl Ip Il Icore Identity Score_EL Rnk_EL Exp BindLevel ------------------------------------------------------------------------------------------------------------ 27 HLA-A*02:01 KLTPLSHEV KLTPLSHEV 0 0 0 0 0 KLTPLSHEV ENSP00000253039 0.9951022 0.009 0.47764 <= SB 59 HLA-A*02:01 KAISGVHTV KAISGVHTV 0 0 0 0 0 KAISGVHTV ENSP00000253039 0.9754907 0.074 0.47764 <= SB 83 HLA-A*02:01 YANAKIYKL YANAKIYKL 0 0 0 0 0 YANAKIYKL ENSP00000253039 0.9058493 0.237 0.47764 <= SB 56 HLA-A*02:01 TVVKAISGV TVVKAISGV 0 0 0 0 0 TVVKAISGV ENSP00000253039 0.7207928 0.635 0.47764 <= WB 57 HLA-A*02:01 VVKAISGVHTV VVISGVHTV 0 2 2 0 0 VVKAISGVHTV ENSP00000253039 0.6767913 0.746 0.47764 <= WB 25 HLA-A*02:01 VTKLTPLSHEV VLTPLSHEV 0 1 2 0 0 VTKLTPLSHEV ENSP00000253039 0.6566866 0.793 0.47764 <= WB 26 HLA-A*02:01 TKLTPLSHEV TLTPLSHEV 0 1 1 0 0 TKLTPLSHEV ENSP00000253039 0.5708980 0.988 0.47764 <= WB 27 HLA-A*02:01 KLTPLSHEVI KLTPLSHEI 0 8 1 0 0 KLTPLSHEVI ENSP00000253039 0.5401132 1.077 0.47764 <= WB 80 HLA-A*02:01 KLGYANAKI KLGYANAKI 0 0 0 0 0 KLGYANAKI ENSP00000253039 0.4893585 1.206 0.47764 <= WB 14 HLA-A*02:01 HLSRQDLTTL HLSQDLTTL 0 3 1 0 0 HLSRQDLTTL ENSP00000253039 0.4232187 1.430 0.47764 <= WB 60 HLA-A*02:01 AISGVHTV AI-SGVHTV 0 0 0 2 1 AISGVHTV ENSP00000253039 0.3835837 1.575 0.47764 <= WB 22 HLA-A*02:01 TLDVTKLTPL TLDVTKLTL 0 8 1 0 0 TLDVTKLTPL ENSP00000253039 0.3285049 1.809 0.47764 <= WB 58 HLA-A*02:01 VKAISGVHTV VAISGVHTV 0 1 1 0 0 VKAISGVHTV ENSP00000253039 0.2115432 2.467 0.47764 55 HLA-A*02:01 STVVKAISGV SVVKAISGV 0 1 1 0 0 STVVKAISGV ENSP00000253039 0.1888403 2.654 0.47764 40 HLA-A*02:01 ATINIGTIGHV ATINIGTHV 0 7 2 0 0 ATINIGTIGHV ENSP00000253039 0.1826325 2.710 0.47764
The prediction output consists of the following columns:
Three amino acid sequences are reported for each peptide (row) in the prediction output:
MAIN REFERENCE
NetMHCpanExp-1.0. The role of antigen expression in shaping the repertoire of HLA presented ligands.
Heli M. Garcia Alvarez 1, Zeynep Koşaloğlu-Yalçın 2, Bjoern Peters2,3, and Morten Nielsen1,4
1
Instituto de Investigaciones Biotecnológicas, Universidad Nacional de San Martín, Buenos Aires, BA 16503, Argentina.
2
Center for Infectious Disease and Vaccine Research, La Jolla Institute for Immunology, La Jolla, California, USA
3
Department of Medicine, University of California, San Diego, La Jolla, California, USA
4
Department of Bio and Health Informatics, Technical University of Denmark, Kgs. Lyngby, DK 28002, Denmark.
In preparation (2022)
OTHER RELEVANT REFERENCES
NNAlign_MA; MHC Peptidome Deconvolution for Accurate MHC Binding Motif Characterization and Improved T-cell Epitope Predictions.
Bruno Alvarez 1, Birkir Reynisson 2, Carolina Barra 1, Søren Buus 3, Nicola Ternette 4, Tim Connelley 5, Massimo Andreatta 1, Morten Nielsen 1,2
1
Instituto de Investigaciones Biotecnológicas, Universidad Nacional de San Martín, Buenos Aires, BA 16503, Argentina.
2
Department of Bio and Health Informatics, Technical University of Denmark, Lyngby, Denmark.
3
Department of Immunology and Microbiology, Faculty of Health Sciences, University of Copenhagen, Denmark.
4
The Jenner Institute, Nuffield Department of Medicine, Oxford, United Kingdom.
5
Roslin Institute, Edinburgh, Midlothian, United Kingdom.
Mol Cell Proteomics (2019); 8(12):2459-2477. DOI: 10.1074/mcp.TIR119.001658. PMID: 31578220
The set of peptides presented on a cell's surface by MHC molecules is known as the immunopeptidome. Current mass spectrometry technologies allow for identification of large peptidomes, and studies have proven these data to be a rich source of information for learning the rules of MHC-mediated antigen presentation. Immunopeptidomes are usually poly-specific, containing multiple sequence motifs matching the MHC molecules expressed in the system under investigation. Motif deconvolution -the process of associating each ligand to its presenting MHC molecule(s)- is therefore a critical and challenging step in the analysis of MS-eluted MHC ligand data. Here, we describe NNAlign_MA, a computational method designed to address this challenge and fully benefit from large, poly-specific data sets of MS-eluted ligands. NNAlign_MA simultaneously performs the tasks of (1) clustering peptides into individual specificities; (2) automatic annotation of each cluster to an MHC molecule; and (3) training of a prediction model covering all MHCs present in the training set. NNAlign_MA was benchmarked on large and diverse data sets, covering class I and class II data. In all cases, the method was demonstrated to outperform state-of-the-art methods, effectively expanding the coverage of alleles for which accurate predictions can be made, resulting in improved identification of both eluted ligands and T-cell epitopes. Given its high flexibility and ease of use, we expect NNAlign_MA to serve as an effective tool to increase our understanding of the rules of MHC antigen presentation and guide the development of novel T-cell-based therapeutics.
Here you will find the datasets used for the independent evaluation of NetMHCpanExp-1.0 method.
MS Ligands
Test_set_I-B_(HPA)(Format: [PEPTIDE] [TARGET_VALUE] [TEST_SET_No] [GENE EXP (TPM)])
Test_set_I-NCI(Format: [PEPTIDE] [TARGET_VALUE] [MHC_ALLELE] [GENE EXP (TPM)])
CD8+ Epitopes
CD8+_epitopes_IEDB(Format: [PEPTIDE] [TARGET_VALUE] [MHC_ALLELE])
Download ALL evaluation data:
Supplementary_Data
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: