DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
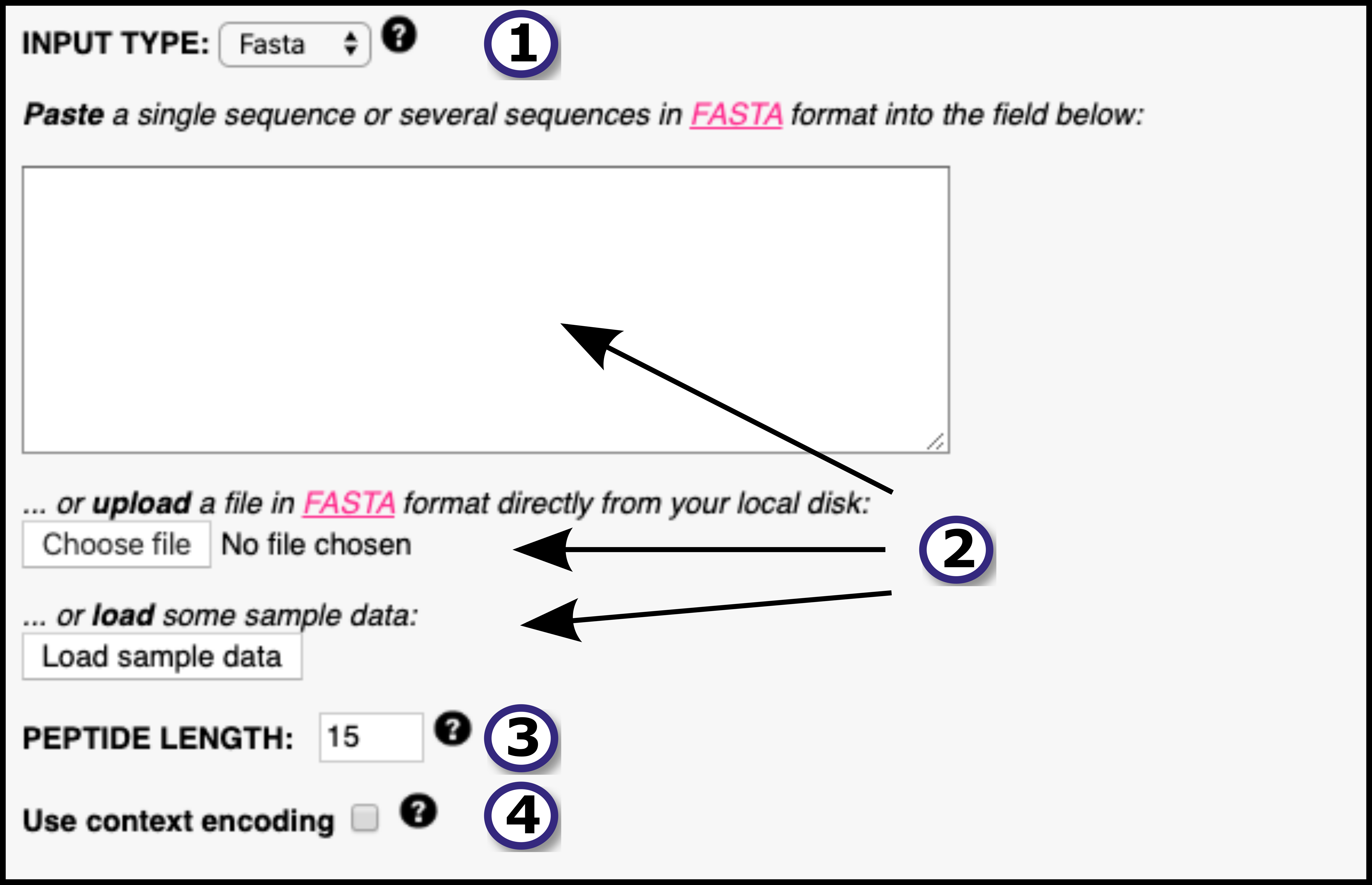
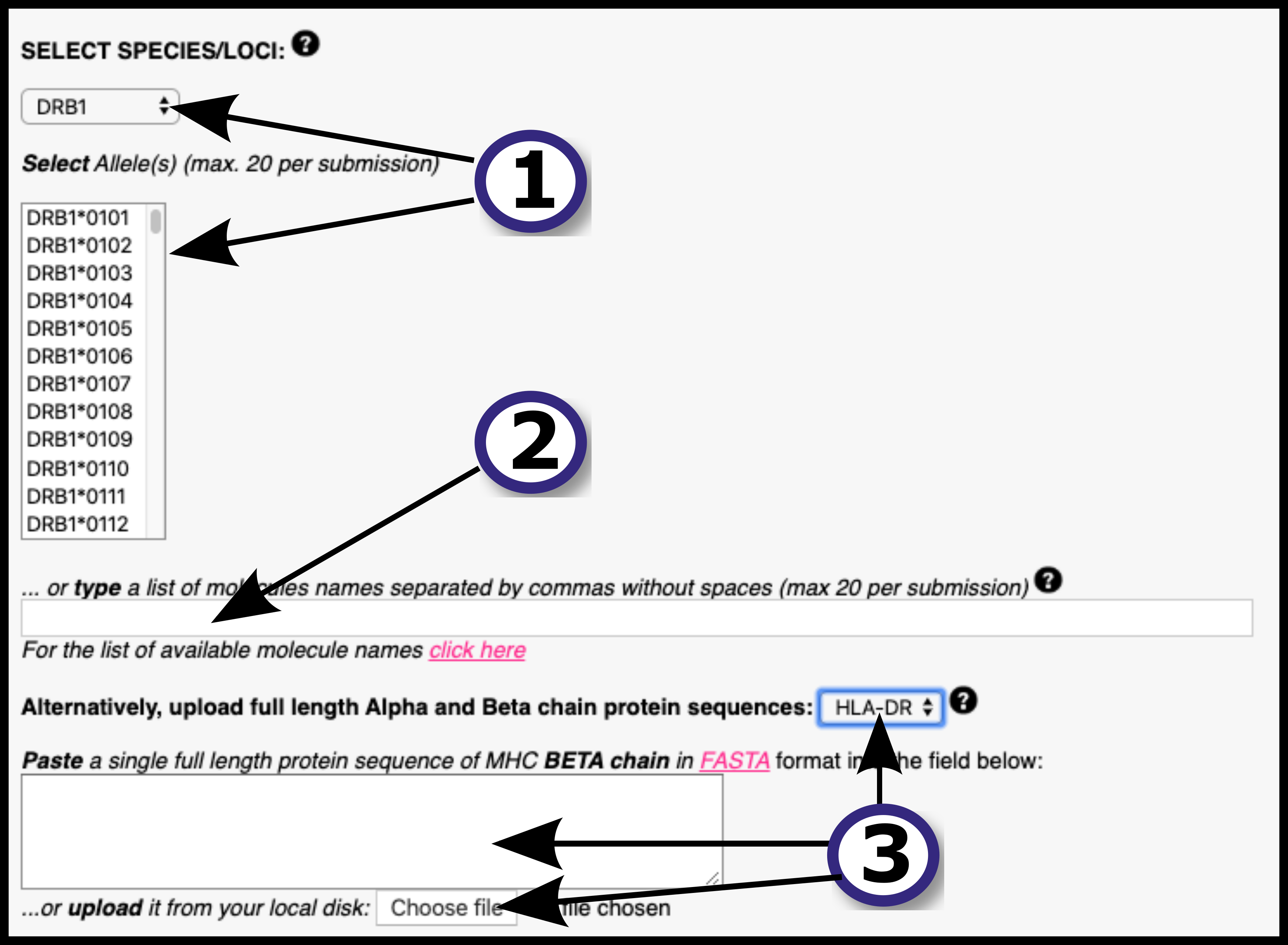
The NetMHCIIpan-4.0 server predicts peptide binding to any MHC II molecule of known sequence using Artificial Neural Networks (ANNs). It is trained on an extensive dataset of over 500.000 measurements of Binding Affinity (BA) and Eluted Ligand mass spectrometry (EL), covering the three human MHC class II isotypes HLA-DR, HLA-DQ, HLA-DP, as well as the mouse molecules (H-2). The introduction of EL data extends the number of MHC II molecules covered, since BA data covers 59 molecules and EL data covers 74. As mentioned, the network can predict for any MHC II of known sequence, which the user can specify as FASTA format. The network can predict for peptides of any length.
The output of the model is a prediction score for the likelihood of a peptide to be naturally presented by and MHC II receptor of choice. The output also includes %rank score, which normalizes prediction score by comparing to prediction of a set of random peptides. Optionally, the model also outputs BA prediction and %rank scores.
New in this version: The two output neuron architechture introduced in NetMHCpan-4.0 permits the inclusion of EL data, and the new training algorithm NNAlign_MA extends training data to ligands of ambiguous allele assignments. The model also, optionally, encodes ligand context.
Note: If you have downloaded the stand alone version of the tool before Maj 1, 2020, please download the data file again from data.tar.gz. The earlier file was missing a few pre-calculated files to estimate percentile rank values.
Refer to the instructions page for more details.
The project is a collaboration between CBS, and LIAI.
View the version history of this server.
For publication of results, please cite:
Benchmark data used to develop this server were obtained from:
NetMHCIIpan 4.0 is available as a stand-alone software package, with the same functionality as the service above. Ready-to-ship packages exist for Linux and MacOSX. There is the tap "download" for academic users; other users are requested to contact CBS Software Package Manager at health-software@dtu.dk.


# NetMHCIIpan version 4.0 # Input is in FASTA format # Peptide length 15 # Prediction Mode: EL # Threshold for Strong binding peptides (%Rank) 2% # Threshold for Weak binding peptides (%Rank) 10% # Allele: DRB1_0101 -------------------------------------------------------------------------------------------------------------------------------------------- Pos MHC Peptide Of Core Core_Rel Identity Score_EL %Rank_EL Exp_Bind BindLevel -------------------------------------------------------------------------------------------------------------------------------------------- 40 DRB1_0101 QGQWRGAAGTAAQAA 3 WRGAAGTAA 1.000 P9WNK5 0.826571 0.45 NA <=SB 39 DRB1_0101 LQGQWRGAAGTAAQA 4 WRGAAGTAA 1.000 P9WNK5 0.729407 0.77 NA <=SB 41 DRB1_0101 GQWRGAAGTAAQAAV 2 WRGAAGTAA 1.000 P9WNK5 0.451999 1.97 NA <=SB 38 DRB1_0101 SLQGQWRGAAGTAAQ 5 WRGAAGTAA 1.000 P9WNK5 0.420826 2.17 NA <=WB 53 DRB1_0101 AAVVRFQEAANKQKQ 3 VRFQEAANK 0.907 P9WNK5 0.096784 6.99 NA <=WB 73 DRB1_0101 STNIRQAGVQYSRAD 3 IRQAGVQYS 1.000 P9WNK5 0.067163 8.66 NA <=WB 26 DRB1_0101 KTQIDQVESTAGSLQ 3 IDQVESTAG 0.993 P9WNK5 0.066535 8.70 NA <=WB 42 DRB1_0101 QWRGAAGTAAQAAVV 1 WRGAAGTAA 0.947 P9WNK5 0.066096 8.73 NA <=WB 52 DRB1_0101 QAAVVRFQEAANKQK 4 VRFQEAANK 0.860 P9WNK5 0.053628 9.80 NA <=WB 14 DRB1_0101 EAGNFERISGDLKTQ 4 FERISGDLK 0.993 P9WNK5 0.044413 10.82 NA 54 DRB1_0101 AVVRFQEAANKQKQE 2 VRFQEAANK 0.573 P9WNK5 0.043962 10.87 NA 72 DRB1_0101 ISTNIRQAGVQYSRA 4 IRQAGVQYS 1.000 P9WNK5 0.038268 11.70 NA
The prediction output for each molecule consists of the following columns:
Major Histocompatibility Complex II (MHC II) molecules play a vital role in the onset and control of cellular immunity. In a highly selective process, MHC II presents peptides derived from exogenousantigens on the surface of antigen-presenting cells for T cell scrutiny. Understanding the rules defining this presentation holds critical insights into the regulation and potential manipulation of the cellular immune system. Here, we apply the NNAlign_MA machine learning framework to analyze and integrate large-scale eluted MHC II ligand mass spectrometry (MS) data sets to advance prediction of CD4+ epitopes. NNAlign_MA allows integration of mixed data types, handling ligands with multiple potential allele annotations, encoding of ligand context, leveraging information between data sets, and has pan-specific power allowing accurate predictions outside the set of molecules included in the training data. Applying this framework, we identified accurate binding motifs of more than 50 MHC class II molecules described by MS data, particularly expanding coverage for DP and DQ beyond that obtained using current MS motif deconvolution techniques. Further, in large-scale benchmarking, the final model termed NetMHCIIpan-4.0, demonstrated improved performance beyond current state-of-the-art predictors for ligand and CD4+ T cell epitope prediction. These results suggest NNAlign_MA and NetMHCIIpan-4.0 are powerful tools for analysis of immunopeptidome MS data, prediction of T cell epitopes and development of personalized immunotherapies.
Here, you will find the data set used for evaluation of NetMHCpan-4.1 and NetMHCIIpan-4.0 methods.
HLA-A02:02
HLA-A02:05
HLA-A02:06
HLA-A02:11
HLA-A11:01
HLA-A23:01
HLA-A25:01
HLA-A26:01
HLA-A30:01
HLA-A30:02
HLA-A32:01
HLA-A33:01
HLA-A66:01
HLA-A68:01
HLA-B07:02
HLA-B08:01
HLA-B14:02
HLA-B15:01
HLA-B15:02
HLA-B15:03
HLA-B15:17
HLA-B18:01
HLA-B35:03
HLA-B37:01
HLA-B38:01
HLA-B40:01
HLA-B40:02
HLA-B45:01
HLA-B46:01
HLA-B53:01
HLA-B58:01
HLA-C03:03
HLA-C05:01
HLA-C07:02
HLA-C08:02
HLA-C12:03
NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data
Submitted 2020.
Please click on the version number to activate the corresponding server.
| 4.0 |
The current server (online since April 2020). New in this version:
|
| 3.2 |
(online since January 2018). New in this version:
|
| 3.1 |
(online since December 2014). New in this version:
|
| 3.0 |
(online since June 2013). New in this version:
|
| 2.1 |
(online since 6 June 2011). New in this version:
|
| 2.0 |
(online since 17 Nov 2010). New in this version:
|
| 1.1 |
(online since 15 April 2010). New in this version:
|
| 1.0 |
Original version (online version until April 15 2010):
Main publication:
|
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: