DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
All the alphabetic symbols not in the allowed alphabet will be converted to X before processing. All the non-alphabetic symbols, including white space and digits, will be ignored.
The sequences can be input in the following two ways:
Both ways can be employed at the same time: all the specified sequences will
be processed. However, there may be not more than 5,000 sequences in one submission. The sequences
may not be longer than 10,000 amino acids.
At any time during the wait you may enter your e-mail address and simply leave the window. Your job will continue; you will be notified by e-mail when it has terminated. The e-mail message will contain the URL under which the results are stored; they will remain on the server for 24 hours for you to collect them.
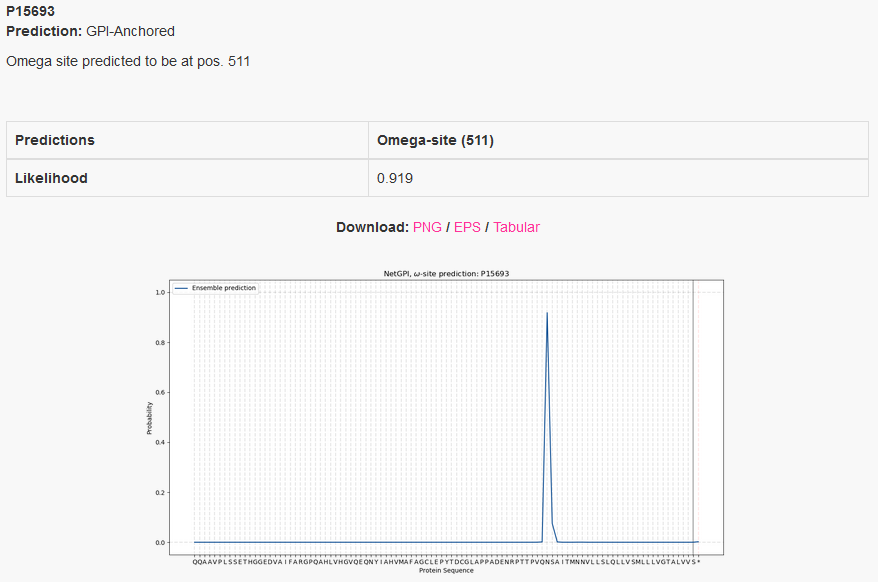
One annotation is attributed to each protein, the one that has the highest probability. If the highest probability is within the amino-acid sequence, then it is considered GPI-anchored and the amino-acid position at the peak is the predicted omega-site. If the highest probability is at the sentinel, here represented by *, then the protein is considered non GPI-anchored.
If a GPI-anchor is predicted, the omega-site position is reported as well.
On the plot we see the likelihood distribution over the protein sequence, with the added sentinel *. Only the last 100 amino-acids are considered.
The datasets for training and benchmarking NetGPI-1.1 can be found here. The dataset is provided in 2-line FASTA format.
The format is as follows:
>uniprot_ac|kingdom|anchoring|pos_from_end|pos_from_beginning|part_no|anchor_exp|omega_exp amino-acid sequence
where:
NetGPI dataset: download
Prediction of GPI-anchored proteins with pointer neural networks
Magnús Halldór Gíslason, Henrik Nielsen, José Juan Almagro Armenteros, Alexander Rosenberg Johansen.
GPI-anchors constitute a very important post-translational modification, linking many proteins to the outer face of the plasma membrane in eukaryotic cells. Since experimental validation of GPI-anchoring signals is slow and costly, computatinal approaches for predicting them from amino acid sequences are needed. However, the most recent GPI predictor is more than a decade old and considerable progress has been made in machine learning since then. We present a new dataset and a novel method, NetGPI, for GPI signal prediction. NetGPI is based on recurrent neural networks, incorporating an attention mechanism that simultaneously detects GPI-anchoring signals and points out the location of their ω-sites. The performance of NetGPI is superior to existing methods with regards to discrimination between GPI-anchored proteins and other secretory proteins and approximate (±1 position) placement of the ω-site.
Current Research in Biotechnology, 3, 6-13, doi: https://doi.org/10.1016/j.crbiot.2021.01.001 (2021)
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: