DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
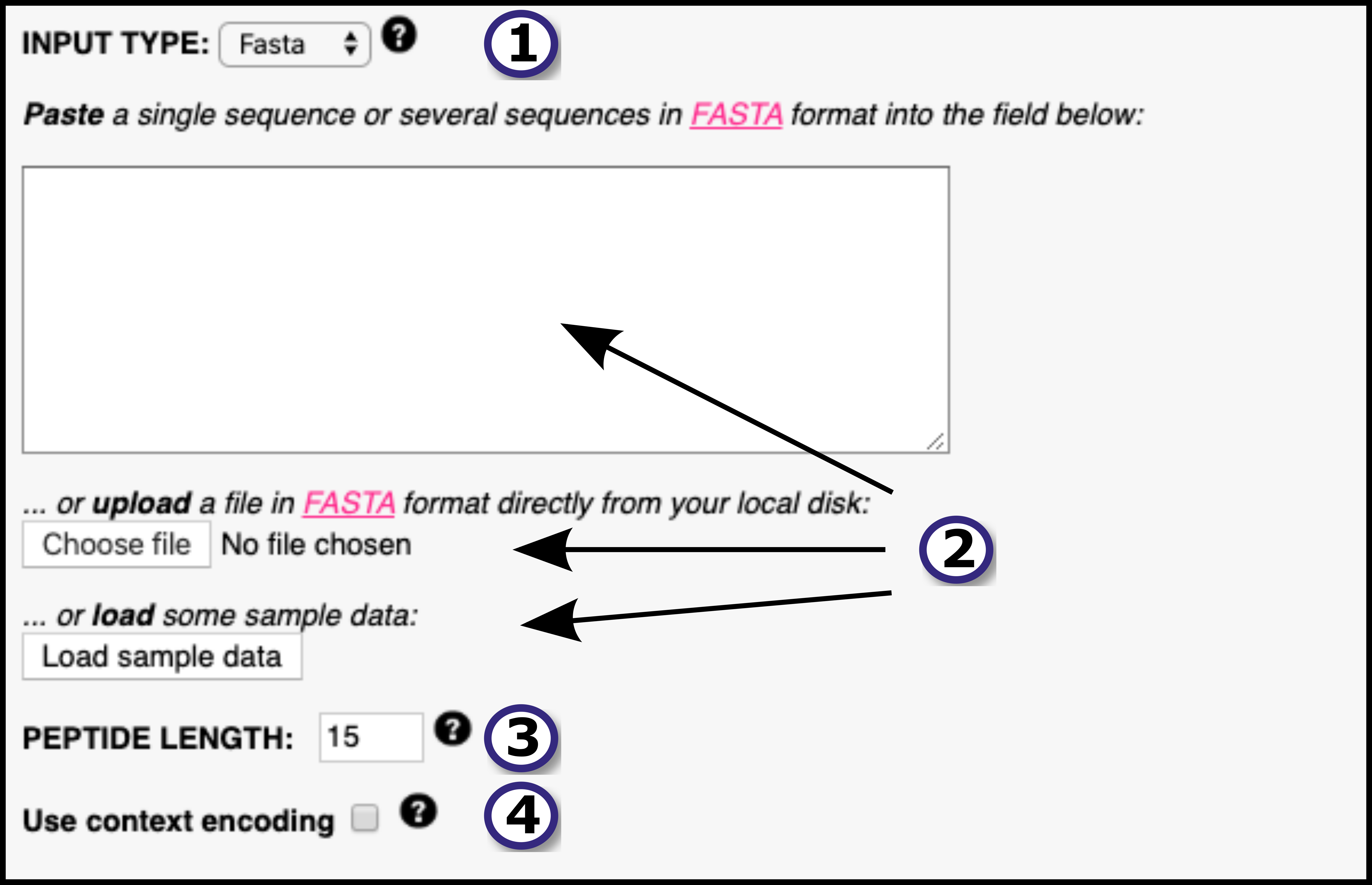
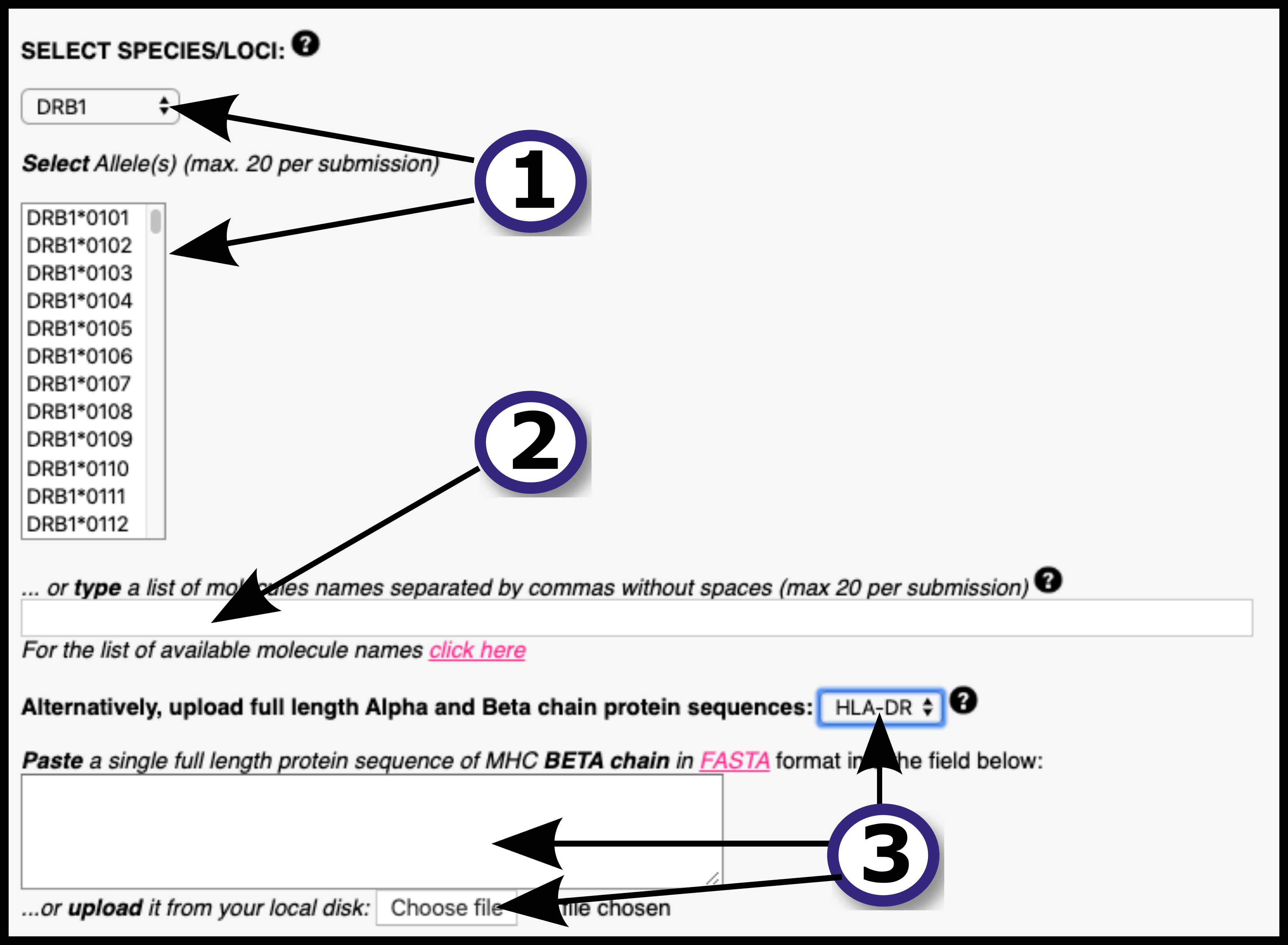
Submission is accepted in two formats - as a list of peptides or as a protein sequence in FASTA format. A comprehensive list of MHC molecules is available for prediction, alternatively the user can upload their MHC protein sequence of interest.
The prediction values are given likelihood for MHC antigen presentation and %Rank score. The percentile rank for a peptide is generated by comparing its score against the scores of 100,000 random natural peptides. For example, if a peptide is assigned a rank of 1%, it means that its predicted affinity is among the top 1% scores for the specified molecule.
Strong and weak binding peptides are identified based on %Rank, with customizable thresholds. You may sort the output based on predicted binding affinity and filter out non-binders.
For publication of results, please cite:


# NetMHCIIpan version 4.0 # Input is in FASTA format # Peptide length 15 # Prediction Mode: EL # Threshold for Strong binding peptides (%Rank) 2% # Threshold for Weak binding peptides (%Rank) 10% # Allele: DRB1_0101 -------------------------------------------------------------------------------------------------------------------------------------------- Pos MHC Peptide Of Core Core_Rel Identity Score_EL %Rank_EL Exp_Bind BindLevel -------------------------------------------------------------------------------------------------------------------------------------------- 40 DRB1_0101 QGQWRGAAGTAAQAA 3 WRGAAGTAA 1.000 P9WNK5 0.826571 0.45 NA <=SB 39 DRB1_0101 LQGQWRGAAGTAAQA 4 WRGAAGTAA 1.000 P9WNK5 0.729407 0.77 NA <=SB 41 DRB1_0101 GQWRGAAGTAAQAAV 2 WRGAAGTAA 1.000 P9WNK5 0.451999 1.97 NA <=SB 38 DRB1_0101 SLQGQWRGAAGTAAQ 5 WRGAAGTAA 1.000 P9WNK5 0.420826 2.17 NA <=WB 53 DRB1_0101 AAVVRFQEAANKQKQ 3 VRFQEAANK 0.907 P9WNK5 0.096784 6.99 NA <=WB 73 DRB1_0101 STNIRQAGVQYSRAD 3 IRQAGVQYS 1.000 P9WNK5 0.067163 8.66 NA <=WB 26 DRB1_0101 KTQIDQVESTAGSLQ 3 IDQVESTAG 0.993 P9WNK5 0.066535 8.70 NA <=WB 42 DRB1_0101 QWRGAAGTAAQAAVV 1 WRGAAGTAA 0.947 P9WNK5 0.066096 8.73 NA <=WB 52 DRB1_0101 QAAVVRFQEAANKQK 4 VRFQEAANK 0.860 P9WNK5 0.053628 9.80 NA <=WB 14 DRB1_0101 EAGNFERISGDLKTQ 4 FERISGDLK 0.993 P9WNK5 0.044413 10.82 NA 54 DRB1_0101 AVVRFQEAANKQKQE 2 VRFQEAANK 0.573 P9WNK5 0.043962 10.87 NA 72 DRB1_0101 ISTNIRQAGVQYSRA 4 IRQAGVQYS 1.000 P9WNK5 0.038268 11.70 NA
The prediction output for each molecule consists of the following columns:
Major histocompatibility complex (MHC) peptide binding and presentation is the most selective event defining the landscape of T cell epitopes. Consequently, understanding the diversity of MHC alleles in a given population and the parametersthat define the set of ligands that can be bound and presented by each of these alleles (the immunopeptidome) has an enormous impact on our capacity to predict and manipulate the potential of protein antigens to elicit functional T cell responses. Liquid chromatography-mass spectrometry (LC-MS) analysis of MHC eluted ligands (EL data) has proven to be a powerful technique for identifying such peptidomes, and methods integrating such data for prediction of antigen presentation have reached a high level of accuracy for both MHC class I and class II. Here, we demonstrate how these techniques and prediction methods can be readily extended to the bovine leukocyte antigen class II DR locus (BoLA-DR). BoLA-DR binding motifs were characterized by EL data derived from cell lines expressing a range of DRB3 alleles prevalent in Holstein-Friesian populations. The model generated (NetBoLAIIpan - available as a web-server at www.cbs.dtu.dk/services/NetBoLAIIpan ) was shown to have unprecedented predictive power to identify known BoLA-DR restricted CD4 epitopes. In summary, the results demonstrate the power of an integrated approach combining advanced MS peptidomics with immunoinformatics for characterization of the BoLA-DR antigen presentation system and provide a novel tool that can be utilised to assist in rational evaluation and selection of bovine CD4 T cell epitopes.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: