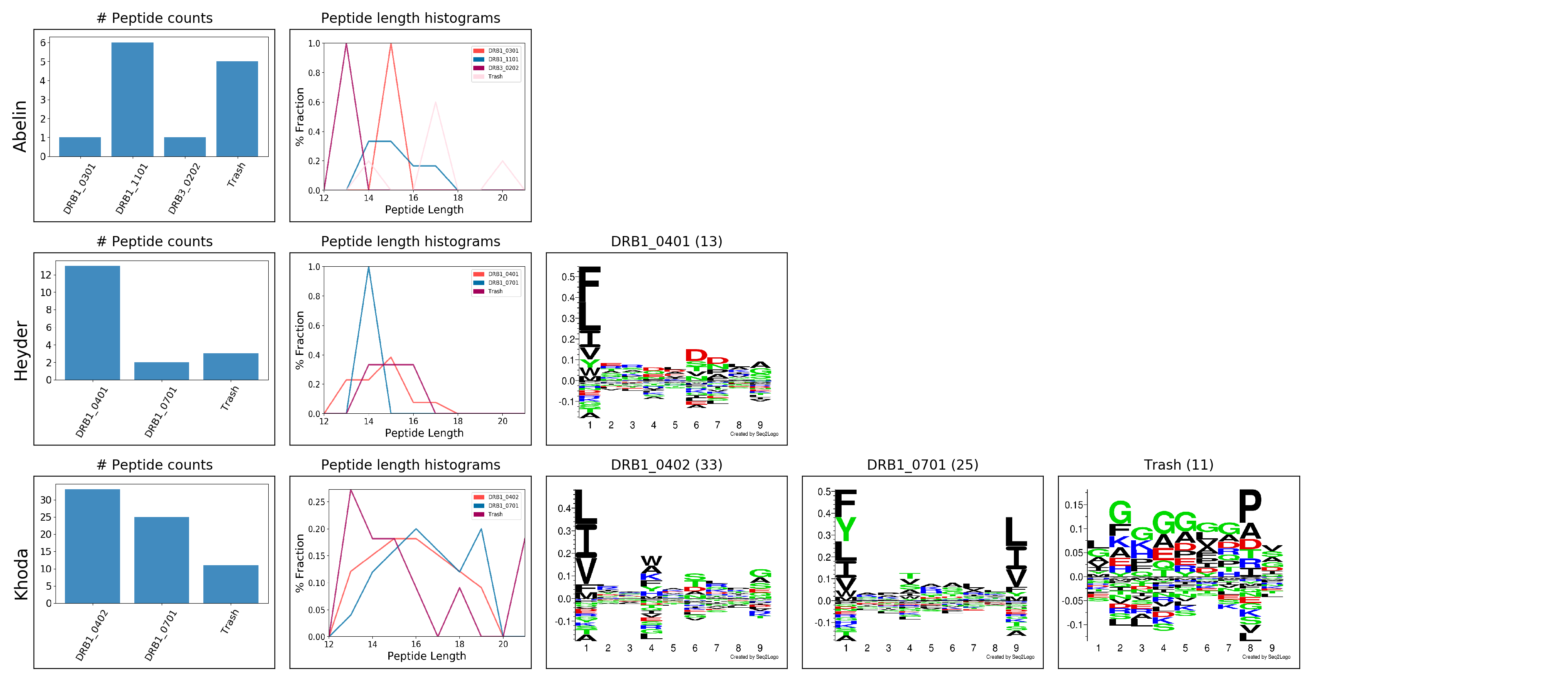
Below is the output from running the MHC class II sample data with default options using MHCMotifDecon-1.0. Note, that the sample data set is very small, and the generated MHC motif and length distributation plots hence are rather noisy.
################################################### # Running MHC_Motif_Decon 1.0 ... # # Call from /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789 # # Input file: /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/file.0 # Number of sequences: 100 # Label-MHC file: /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/file.1 # MHC class: II # Length range: 12-21 # RANK threshold: 20 # Minimum quantity of sequences for logo plotting: 10 # MHC counts plots will be included. # MHC length histograms will be included. # Run ID: run_21019 # Dirty mode enabled. # # Creating output folder /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019... # DONE. # # Deconvoluting Peptide-MHCs... # 100/100 # Deconvolution DONE. # # Reading alleles for Abelin... # Reading alleles for Heyder... # Reading alleles for Khoda... # WARNING! Low quantity of sequences (1) found for DRB1_0301 in Abelin. This allele will not be plotted. WARNING! Low quantity of sequences (6) found for DRB1_1101 in Abelin. This allele will not be plotted. WARNING! Low quantity of sequences (1) found for DRB3_0202 in Abelin. This allele will not be plotted. WARNING! Low quantity of sequences (5) found for Trash in Abelin. This allele will not be plotted. # # Found 13 sequences for DRB1_0401 in Heyder WARNING! Low quantity of sequences (2) found for DRB1_0701 in Heyder. This allele will not be plotted. WARNING! Low quantity of sequences (3) found for Trash in Heyder. This allele will not be plotted. # # Found 33 sequences for DRB1_0402 in Khoda # Found 25 sequences for DRB1_0701 in Khoda # Found 11 sequences for Trash in Khoda # # WARNING! The logo for DRB1_0301 in Abelin will not be generated. # WARNING! The logo for DRB1_1101 in Abelin will not be generated. # WARNING! The logo for DRB3_0202 in Abelin will not be generated. # WARNING! The logo for Trash in Abelin will not be generated. # 1/4 Making logo for DRB1_0401 in Heyder... # WARNING! The logo for DRB1_0701 in Heyder will not be generated. # WARNING! The logo for Trash in Heyder will not be generated. # 2/4 Making logo for DRB1_0402 in Khoda... # 3/4 Making logo for DRB1_0701 in Khoda... # 4/4 Making logo for Trash in Khoda... # # Generating peptide count histograms... DONE. # # Generating peptide length histograms... DONE. # # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Abelin_count_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Abelin_peptide_length_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Heyder_count_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Heyder_peptide_length_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos/Heyder@DRB1_0401.logo-001.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Khoda_count_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/data/Khoda_peptide_length_histogram.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos/Khoda@DRB1_0402.logo-001.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos/Khoda@DRB1_0701.logo-001.png # Plotting /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos/Khoda@Trash.logo-001.png # # SAVING LOGOS PLOT... # DPI: 200.0 # PDF: /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos.pdf # PNG: /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019/logos.png # # MHC_Motif_Decon has finished. # # Results are stored in /var/www/webface/tmp/server/mhcmotifdecon-1.0/61DCAF1F000013A1AED09789/run_21019 ################################################### logos/ logos/Heyder@DRB1_0401.logo.eps logos/Heyder@DRB1_0401.logo.txt logos/Heyder@DRB1_0401.logo_freq.mat logos/Heyder@DRB1_0401.logo-001.png logos/Khoda@DRB1_0402.logo.eps logos/Khoda@DRB1_0402.logo.txt logos/Khoda@DRB1_0402.logo_freq.mat logos/Khoda@DRB1_0402.logo-001.png logos/Khoda@DRB1_0701.logo.eps logos/Khoda@DRB1_0701.logo.txt logos/Khoda@DRB1_0701.logo_freq.mat logos/Khoda@DRB1_0701.logo-001.png logos/Khoda@Trash.logo.eps logos/Khoda@Trash.logo.txt logos/Khoda@Trash.logo_freq.mat logos/Khoda@Trash.logo-001.png logos/Abelin_count_histogram.png logos/Abelin_peptide_length_histogram.png logos/Heyder_count_histogram.png logos/Heyder_peptide_length_histogram.png logos/Khoda_count_histogram.png logos/Khoda_peptide_length_histogram.png
Motif Deconvolution plot:
Link to prediction file Output_file.xls
Link to tar.gz file with logo/image files data.tar.gz