DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
The LYRA server predicts structures for either T-Cell Receptors (TCR) or B-Cell Receptors (BCR) using homology modelling. Framework templates are selected based on BLOSUM score, and complementary determining regions (CDR) are then selected if needed based on a canonical structure model and grafted onto the framework templates.
Input a pair of TCR or BCR protein sequences in any order. One sequence per field, no fasta headers. After submission of sequences is complete, it is possible to manually select new templates and resubmit. Prediction of structure should take less than a few minutes depending on queue status.
Database version: 2015-04-13
The LYRA prediction server requires a pair of TCR chains or BCR chains and allows both protein sequences and nucleic acid sequences.
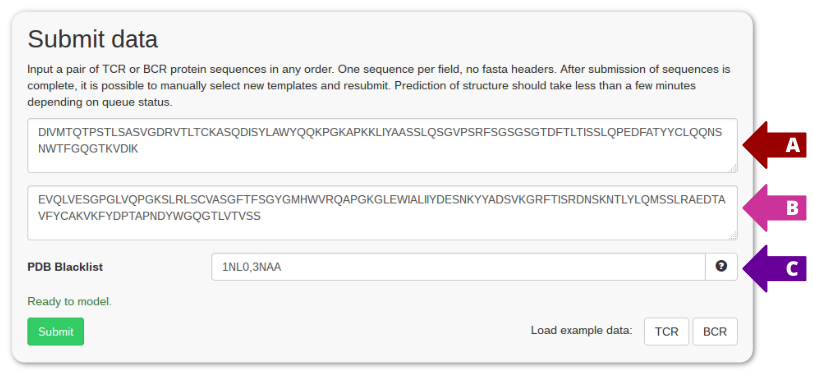
Paste chain sequences into field 1 and 2 as marked by arrows A and B above. If any specific PDB templates should be ignored when modelling, use the optional blacklist field (usually used for benchmarking) marked by arrow C in the screenshot above. When all required fields are filled click the submit button to get your results.
After the server successfully finishes the job, a summary page shows up. If an error happens during modelling a log will appear specifying the error.
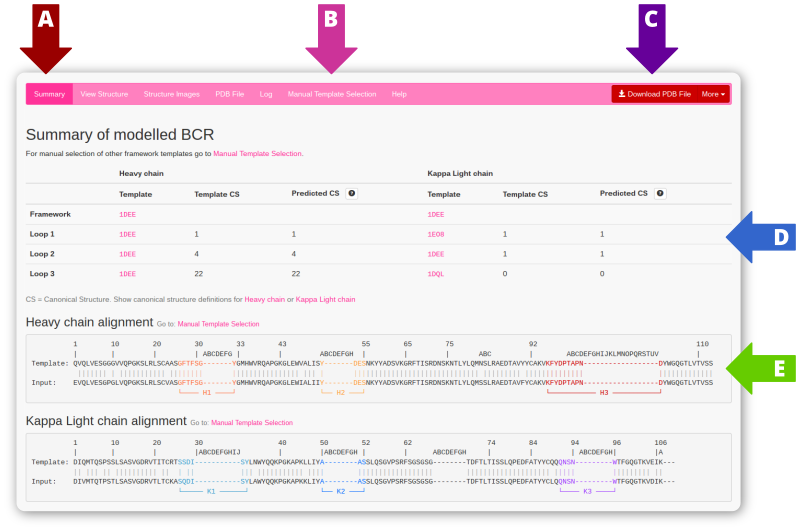
Use the navigation bar (arrow A) to flip through the various output pages. The default page is the summary, which shows a table of template structures and calculated canonical structures for both chains (arrow D) as well as the aligned sequences (arrow E) with the input sequence, the template sequence and the variable loops highlighted. Use the download button (arrow C) to download the generated PDB file, or select "more" to get alternate download options.
After results are generated an option of manually changing the framework templates for each chain is revealed (arrow B above).
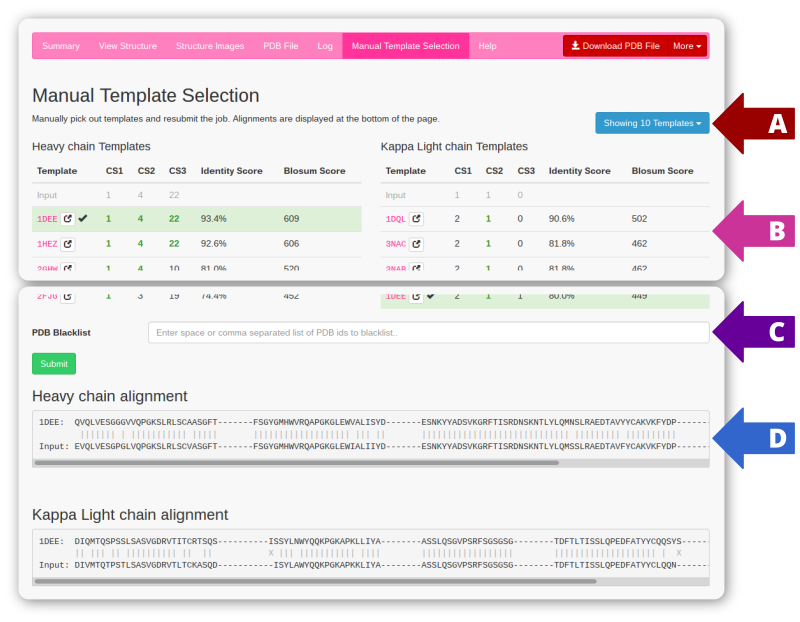
On the manual template selection page the top 10 scoring templates are shown in a table (arrow B). The number of shown templates can be changed by selecting a different number using the dropdown at arrow A.
When entering the Manual Template Selection tab the current framework templates are highlighted within the top templates. Change to the wanted framework templates and click the submit button. Whenever a new template is selected, the alignment is automatically updated to show the template:input sequence alignment (arrow D).
This creates and new result page with the new selected framework templates.
Note that only the framework templates are changed. The LYRA server will still select the optimal canonical structures for each CDR loop with a different length or canonical structure. Specific structure templates can be blacklisted using the blacklist input field (arrow C above).
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: