DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
The program is based on a large dataset of haplotype frequencies obtained by DNA-typing 2.9 million individuals divided in 21 detailed ethnicity categories and 6.59 million individuals divided in 5 broad ethnicity categories. These data are retrieved from the voluntary bone marrow donor program Be The Match Registry made by The National Marrow Donor Program (NMDP) [23806270].
By defining a threshold or top, the output is restricted to a certain probability measure or the top highest probabilities. If top highest probabilities is chosen, only the associated alleles with a probability of 5% or more, will be included in the output. The server will take either donor or population data as input. Donor data will be a file containing donor sample IDs together with the DRB1 found in each donor, while the population data will be a file with DRB1 together with the frequency of finding that specific allele in the population.
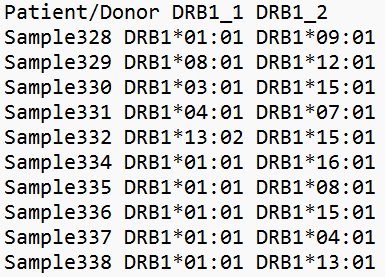 Example of donor input Download example file |
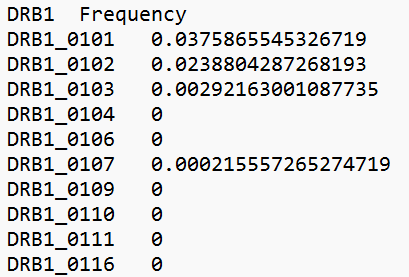 Example of population input Download example file |
Output:
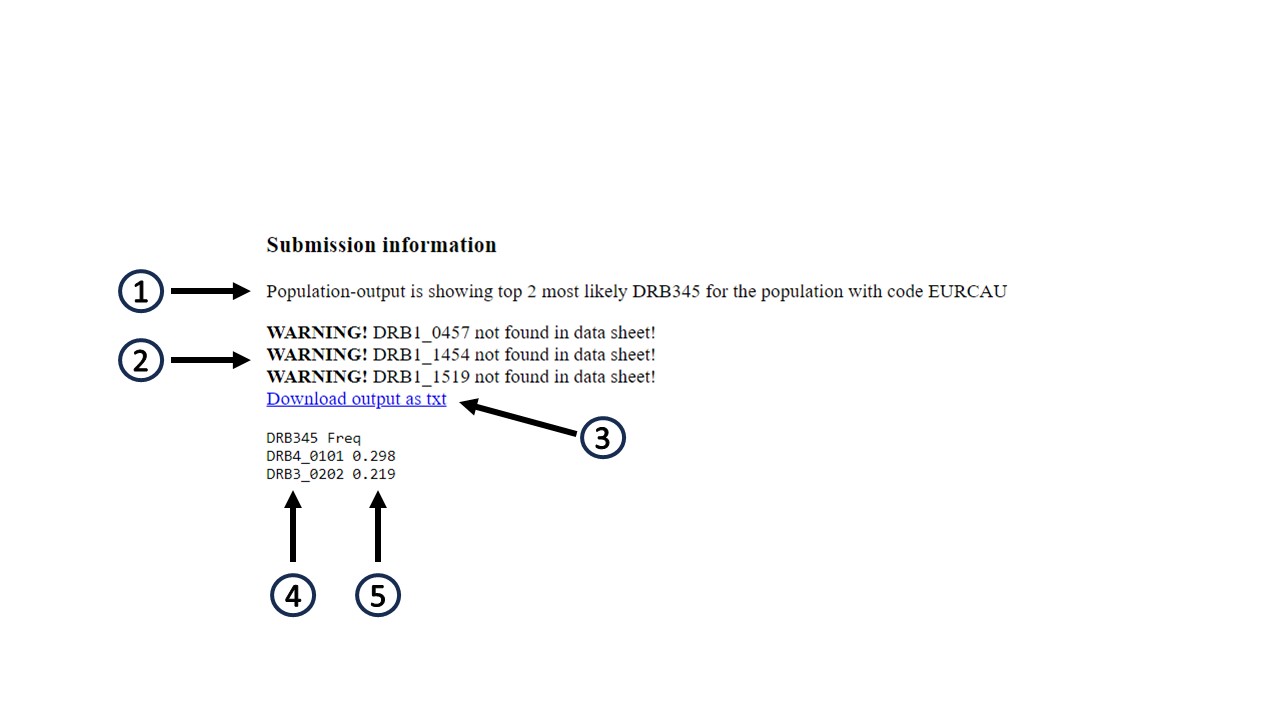
The output format depends on the input data.
For input data of donor type, see figure: 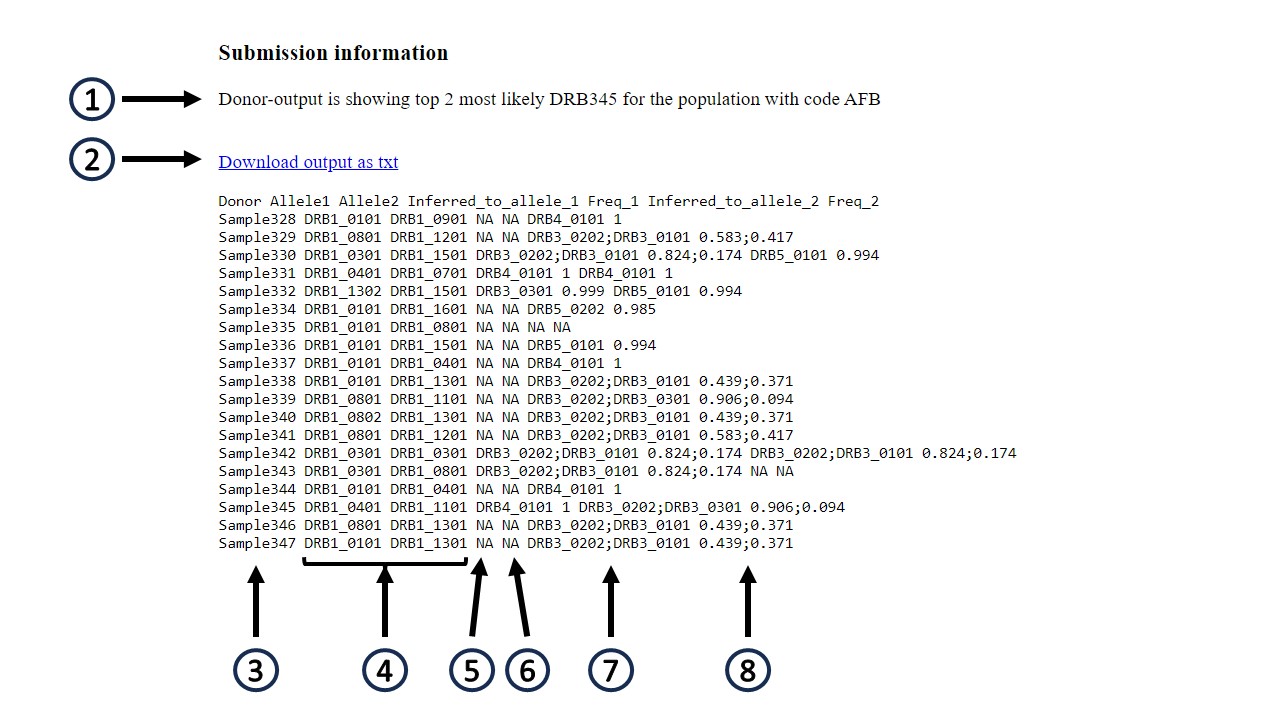
Insights into peptide binding to HLA class II molecules is essential when studying the biological mechanisms
behind cellular immunity, autoimmune diseases, and the development of immunotherapies and peptide vaccines.
Currently, most of the publicly available data used to train state-of-the-art binding prediction methods for
HLA-DR only includes DRB1 information. The role of the paralogue alleles, HLA-DRB3/4/5, and their strong
linkage disequilibrium to DRB1 is often omitted when typing HLA-II alleles. This leads to ambiguities when
making disease associations and interpreting HLA-restricted immune data. To resolve this issue, we present
HLAAssoc-1.0, a method to infer HLA-DRB3/4/5 alleles by linkage disequilibrium to HLA-DRB1.
We illustrate the usage of the tool and the importance of the integration of HLA-DRB3/4/5 alleles in the
data analysis in different case studies including the interpretation of immunopetidomics data.
Additionally, we infer allele information for the data used for training of NetMHCIIpan lacking
HLA-DRB3/4/5 allele information and demonstrate that the retrained method achieved improved performance.
In all cases, inferring HLA-DRB3/4/5 allele presence in non-fully typed HLA-II assays resulted in improved
allele and motif deconvolutions.
HLAAssoc-1.0 is available at https://services.healthtech.dtu.dk/services/HLAAssoc-1.0/
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: