1. Purpose and data sources
The HExpoChem server is an integrative database containing diverse
chemicals in the aim to explore human health risk from chemical exposure. The
main concept of HExpoChem is to aggregate bioactivities (of proteins)
associated to chemicals found in five major sources, namely in drugs, cosmetics,
foods, industrial chemicals and that might interfere with human metabolites. From
the chemical-protein interactions, generation of protein complexes was performed (protein-protein
interactions network and protein-protein associations network) to propose potential biological
outcomes (diseases, pathways, GO terms) associated to protein perturbation by
chemicals.
The sources of the chemical information integrated in HExpoChem are described
below:
-
The drugs source contains all small molecules approved by the U.S. Food and Drug
Administration (FDA) as well as withdrawn drugs.
-
The cosmetics source include all chemicals known to be present in personal
care product such as antipersistant, hair spray, makeup, shampoo.
-
The foods source is a compilation of small molecules known to naturally occur in
food (like phenolic compounds or flavors) and substances added to food
including substances regulated by the FDA as direct, "secondary" direct and
color additives, and Generally Recognized As Safe (GRAS).
-
The industrial chemicals source represents small molecules that are present in
our environment. They include pesticides, chemicals found in home maintenance
products (for example solvent in paint), in home office and plasticizers.
-
The metabolites source contains all human small molecules identified in the human
metabolism, as described in the BiGG database.
Compounds are not source specific, i.e. they can be present in several sources.
For example, human can be exposed to butylparaben via the foods (as additive to
beer to delay microbial growth), the cosmetics (as preservative in eye shadow,
baby lotion...) and industrial chemicals (via indoor air or dust produced by
aerosolized products).
2. Submission
The user will have the possibility to search for proteins annotated to a
specific chemical (chemical search) or biological information associated to a specific protein
and a complex of proteins (protein search).
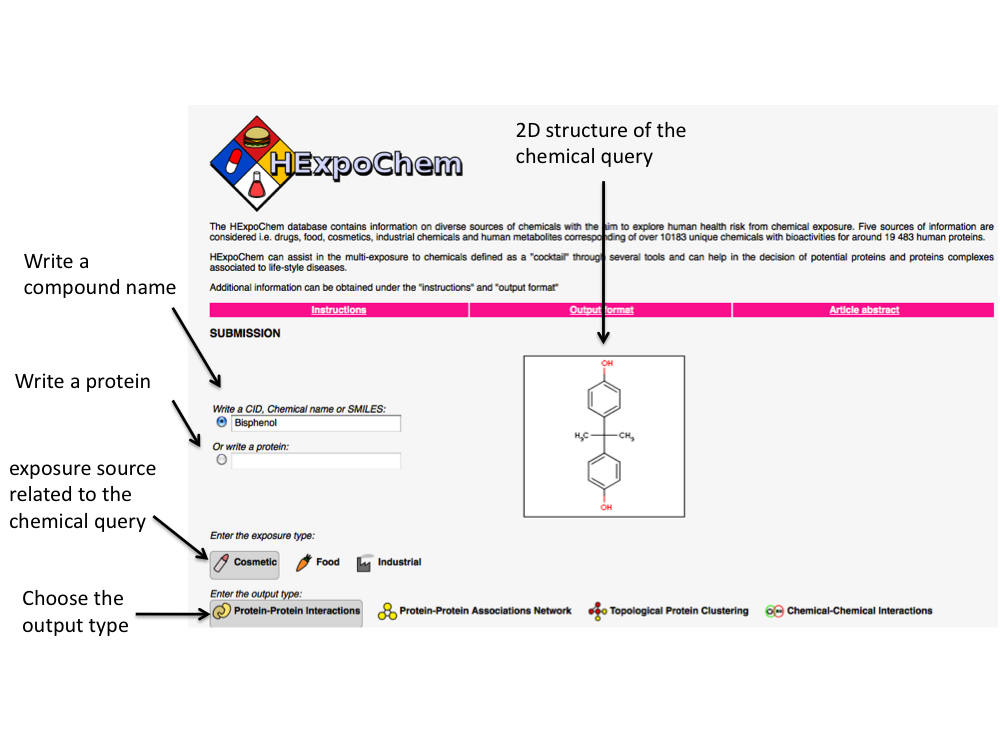
2.1 Chemical search (Write a CID, chemical name or SMILE)
There are several ways to search for a chemical. The user can:
-
Write a chemical name in the box (for example bisphenol A). Synonyms can be
used as well.
-
Write a SMILES code (a SMILES is a string that encode the chemical structure of
a compound). In the case of bisphenol A, the canonical SMILES is
CC(C)(C1=CC=C(C=C1)O)C2=CC=C(C=C2)O
-
Write a CID code (PubChem ID code). PubChem is one of the largest public
available repositories of small molecules and compounds are identified with a
CID code. For bisphenol A, the user will have to enter CID000006623)
Once a chemical has been defined, it will be depicted automatically in which
exposure source it can be found and how many proteins it interacts with. For example, bisphenol A might be present in
Cosmetic, Food and Industrialand interacts with 832 proteins.
2.2 Biological output
The user has the possibility to select between four different outputs associated
to the chemical.
-
Protein-protein interactions (PPI) will allow visualizing the proteins perturbed by
the query chemical and their involvement in a proteins complex. Direct
protein interactions to the protein input are considered. Enriched biological
annotations are then linked to each proteins complex
-
Protein-protein associations network (PPAN) is a predictive method based on the assumption
that if a chemical affect two proteins, then both proteins are
deemed associating in chemical space. Here, we considered only direct protein
neighbors for each protein perturbed by a chemical. Enriched biological
annotations are then linked to each PPAN developed.
-
Topological protein clustering is an alternative to the protein-protein
associations network where a graph clustering algorithm, the Markov cluster algorithm
(MCL), was applied on the entire biological network in order to group proteins
by their connection density. Enriched biological annotations are then linked to each
cluster.
-
Chemical-Chemical Interactions (CCI) is based on the assumption that two chemicals
bioactive for the same set of proteins can have a cumulative effect on these
proteins and thus increase the biological effects. The CCI approach was
implemented in three conditions. For a query compound, search for agonist-antagonist (antagonism effect);
agonist-agonist (synergistic effect) and antagonist-antagonist (synergistic effect)
Enriched biological annotations are then linked to each set of proteins
interacting for two chemicals.
NB: About the protein-protein associations network (PPAN and the topological
protein clustering (TPC),
the user will have to choose the exposure type (if more than one is
depicted with the chemical search). As, PPAN and TPC are dependent of the
chemical-protein interactions, the development of a network for each type of
exposures will limit biais from drug-target interactions, that have been much
more investigated than the other sources.
2.3 Protein search
The user can search directly to proteins that are perturbed by chemicals and
find relevant biological information connected to it or to protein complexes in
which the input protein is involved to. To do so, the user has to write a gene
name. For example, to find information for the estrogen receptor, ESR1 has to be
written (which is the gene name identifier of this receptor). It is possible to
find such gene names at the uniprot server (www.uniprot.org).
3. Submit the job
Click on the
"Submit" button. The status of your job (either 'queued'
or 'running') will be displayed and constantly updated until it terminates and
the server output appears in the browser window.
At any time you may enter your e-mail address and simply leave
the window. Your job will continue and you will be notified by e-mail when the
job is finished. The e-mail message will contain the URL under which the results are
stored; they will remain on the server for 24 hours for you to collect them.