DiscoTope-3.0 predicts per-residue epitope propensity of input protein structures. The server requires input protein structures in the PDB format. These may either be uploaded by the user or downloaded by the DiscoTope-3.0 server.
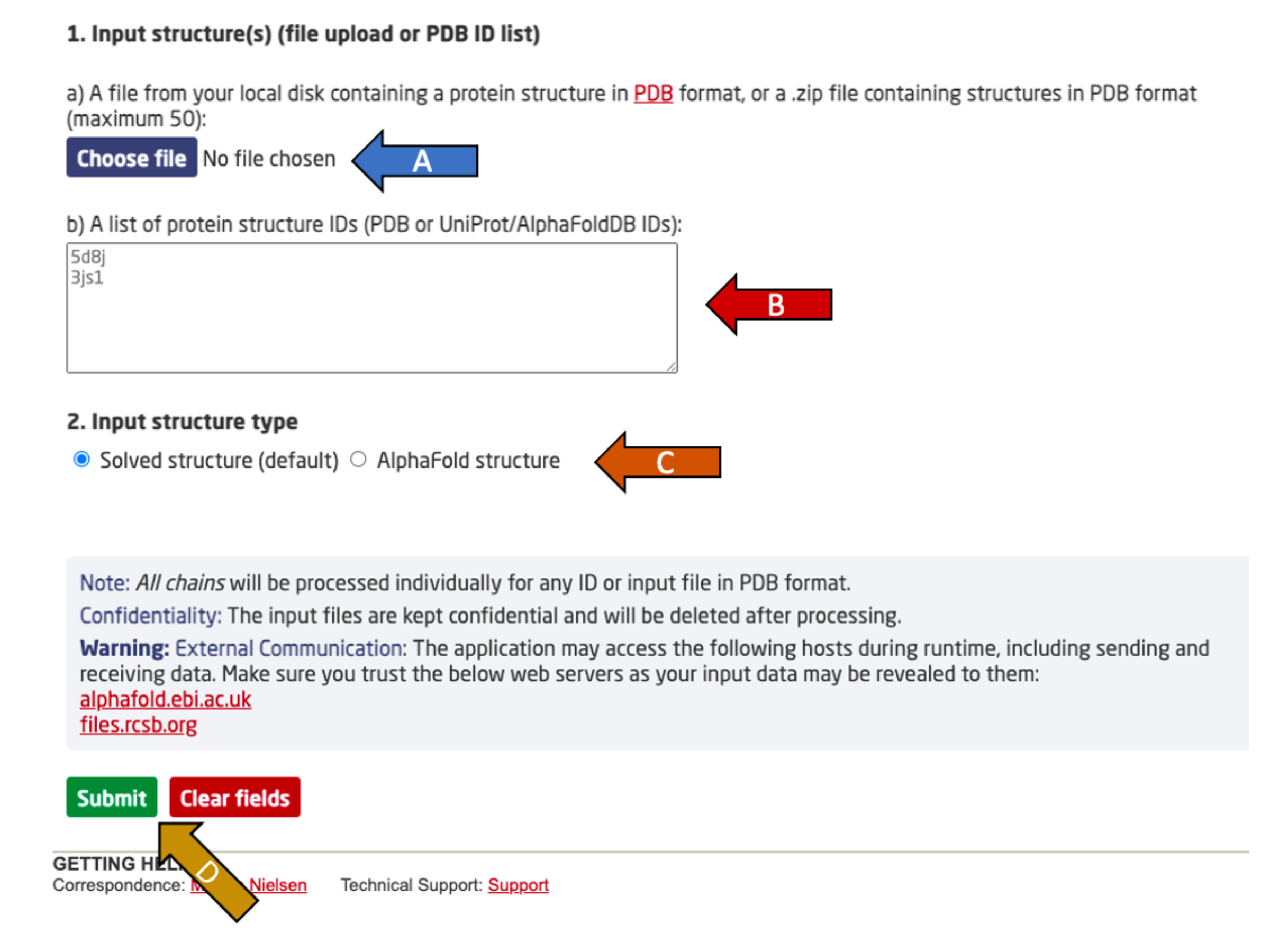
Choose between either:
- Upload a protein PDB structure or a compressed ZIP archive with the Choose file button (arrow A). The ZIP file may contain up to 50 PDBs. The chosen file may not be larger than 30 MB.
- OR
- Enter a list of protein structure IDs, to download from either RCSB or AlphaFoldDB based on the flag set in arrow C. No chain specification should be used (e.g. 5d8j_A). Up to 50 IDs may be given, with one ID per line.
After providing one of the two input options, specify the input structure type (arrow C). Experimental PDB structures (solved) and AlphaFold2 predicted structures are supported.
The suggested calibrated score thresholds correspond to observed epitope percentile scores in the validation set, matching expected recall rates at the given thresholds (see paper).
- Higher confidence (1.50, recall up to ~30 %)
- Moderate confidence (0.90, recall up to ~50 %, default)
- Lower confidence (0.40, recall up to ~70 %)
Finally, click submit to complete the submission (arrow D).