Output format
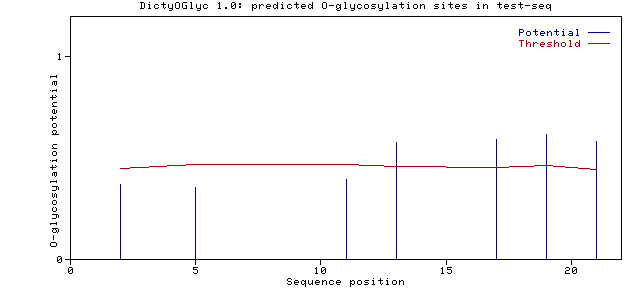
If the Potential > Threshold then O-glycosylation is predicted at that
site. Predicted sites are assigned 'G'.
In this example Thr 13, 17, 19 and 21 are predicted to be O-glycosylated.
The higher the potential the higher is the confidence of the prediction.
The Threshold takes into account surface/buried predictions
of the particular site, and indicates
a value above which sites are predicted to be on the surface
and O-glycosylated.
The figure (using: Generate graph) shows the potential and threshold
versus sequence position.
Alternative non-predicted sites may be found where the potential is
high (>0.4) and threshold low (<0.55).
Name: test_seq Length: 22
GSDWSGVCKITATPAPTVTPTV <-- Submitted sequence.
............G...G.G.G. <-- The assignments. G stands for predicted
GlcNAc O-glycosylation at site.
SeqName Residue Number Potential Threshold Assignment
test_seq Ser 0002 0.3727 0.4493 .
test_seq Ser 0005 0.3553 0.4687 .
test_seq Thr 0011 0.3973 0.4687 .
test_seq Thr 0013 0.5762 0.4606 G
test_seq Thr 0017 0.5945 0.4567 G
test_seq Thr 0019 0.6180 0.4638 G
test_seq Thr 0021 0.5827 0.4455 G
Graphics in
PostScript
