DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
 |
 |
|
Restrictions:
At most 50 sequences and 70,000 amino acids per submission;
each sequence not more than 4,000 amino acids.
Note that the first 10 characters (after '>') in the fasta header line must be unique.
Confidentiality:
The sequences are kept confidential and will be deleted
after processing.
CITATIONS
For publication of results, please cite:
Scanning the available Dictyostelium discoideum proteome for
O-linked GlcNAc glycosylation sites using neural networks.
R. Gupta, E. Jung, A.A. Gooley, K.L. Williams, S. Brunak and J. Hansen.
Glycobiology: 9(10):1009-22, 1999.
View the abstract.
O-GlycBase
See the O-GlycBase database for a revised database of O- and C-glycosylated proteins.
In order to use the DictyOglyc server for prediction on amino-acids sequences either:
>seq1
ASTPGHTIIYEAVCLHNDRTTIP
>seq2
ASQKRPSQRHGSKYLATASTMDHARHGFLPRHRDTGILDSIGRFFGGDRGAPK
NMYKDSHHPARTAHYGSLPQKSHGRTQDENPVVHFFKNIVTPRTPPPSQGKGR
KSAHKGFKGVDAQGTLSKIFKLGGRDSRSGSPMARR
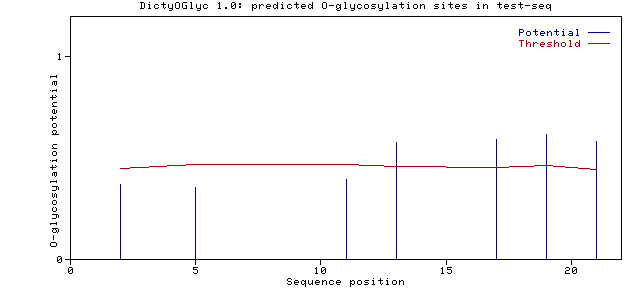
Name: test_seq Length: 22 GSDWSGVCKITATPAPTVTPTV <-- Submitted sequence. ............G...G.G.G. <-- The assignments. G stands for predicted GlcNAc O-glycosylation at site. SeqName Residue Number Potential Threshold Assignment test_seq Ser 0002 0.3727 0.4493 . test_seq Ser 0005 0.3553 0.4687 . test_seq Thr 0011 0.3973 0.4687 . test_seq Thr 0013 0.5762 0.4606 G test_seq Thr 0017 0.5945 0.4567 G test_seq Thr 0019 0.6180 0.4638 G test_seq Thr 0021 0.5827 0.4455 G
We expect the server to predict sites more 'specifically', that is to pick upO-GlcNAc sites fromsecreted and membrane proteins of D.discoideum. Predictions with the new server may vary from those made with DictyOGlyc v/1.0. On a general basis, the server picks fewer glycosylation sites presumably excluding more false positives.
Feedback, Comments and suggestions are most welcome.
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: