DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DeepLocPro predicts the subcellular localization of prokaryotic proteins. It can differentiate between 6 different localizations: Cytoplasm, Cytoplasmic membrane, Periplasm, Outer membrane, Cell wall and surface, and extracellular space.
New book chapter: Practical Applications of Language Models in Protein Sorting Prediction: SignalP 6.0, DeepLoc 2.1, and DeepLocPro 1.0, pp. 153-175 of Dukka B. KC (ed.): Large Language Models (LLMs) in Protein Bioinformatics (MIMB, volume 2941), 2025
Eukaryotic proteins: To predict the locations of proteins in eukaryotes, use DeepLoc.
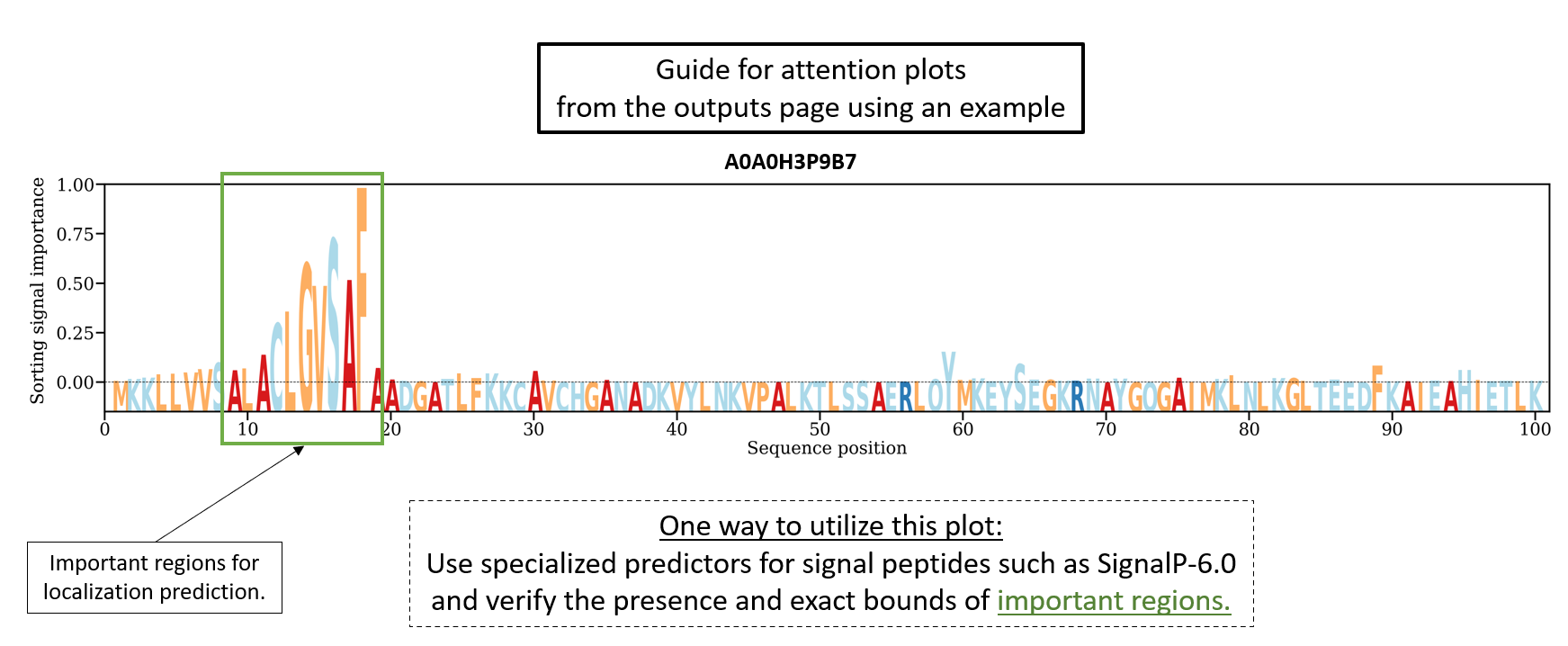
The DeepLocPro 1.0 server predicts the subcellular localization of prokaryotic proteins using a neural network based algorithm trained on Uniprot and ePSORTdb proteins with experimental evidence of subcellular localization. It only uses the sequence information to perform the prediction. The importance of each amino acid in the predicted localization is also included as an "attention" plot. Positions in the sequence with a high attention value are deemed more relevant for the prediction. This does not mean that a particular amino acid is very important for the prediction but that a region in the neighbourhood of those positions has more weight in the final prediction of the model.
The DeepLocPro 1.0 server also accepts the organism group of the input sequences as input.
The DeepLocPro 1.0 server requires protein sequence(s) in fasta format, and can not handle nucleic acid sequences.
Two different versions of the output can be selected before running DeepLocPro 1.0. The long output will generate an attention plot per sequence while the short output will not generate any plots.
Paste protein sequence(s) in fasta format or upload a fasta file.
After the server successfully finishes the job, a summary page shows up. If an error happens during the prediction a log will appear specifying the error.
The DeepLocPro output is composed of three main components:
The dataset used to train and test the DeepLocPro 1.0 server is available here:
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: