DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
Paste or upload protein sequence(s) as fasta format to predict potential B-cell epitopes. Prediction can take a few minutes per sequence.
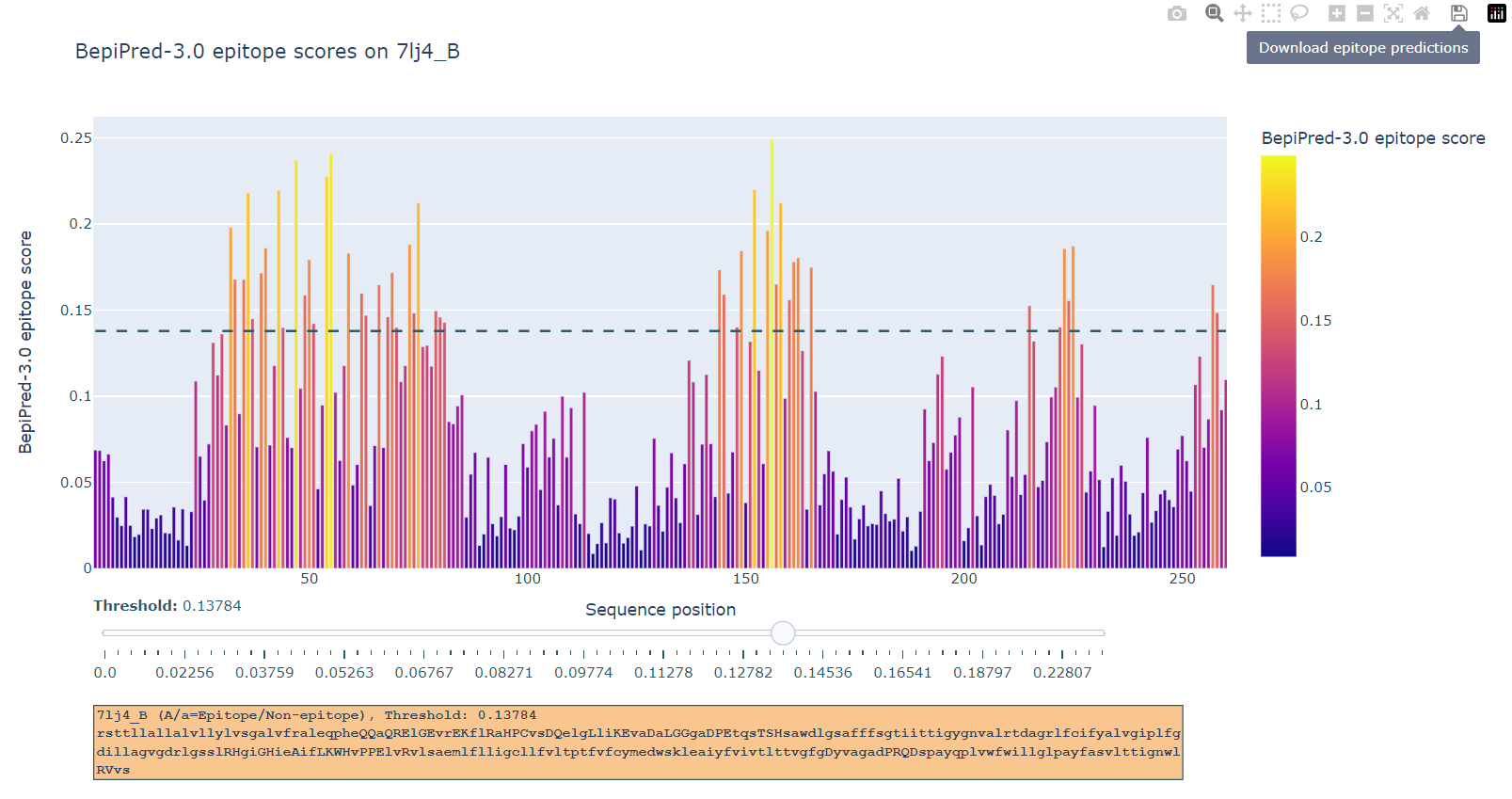
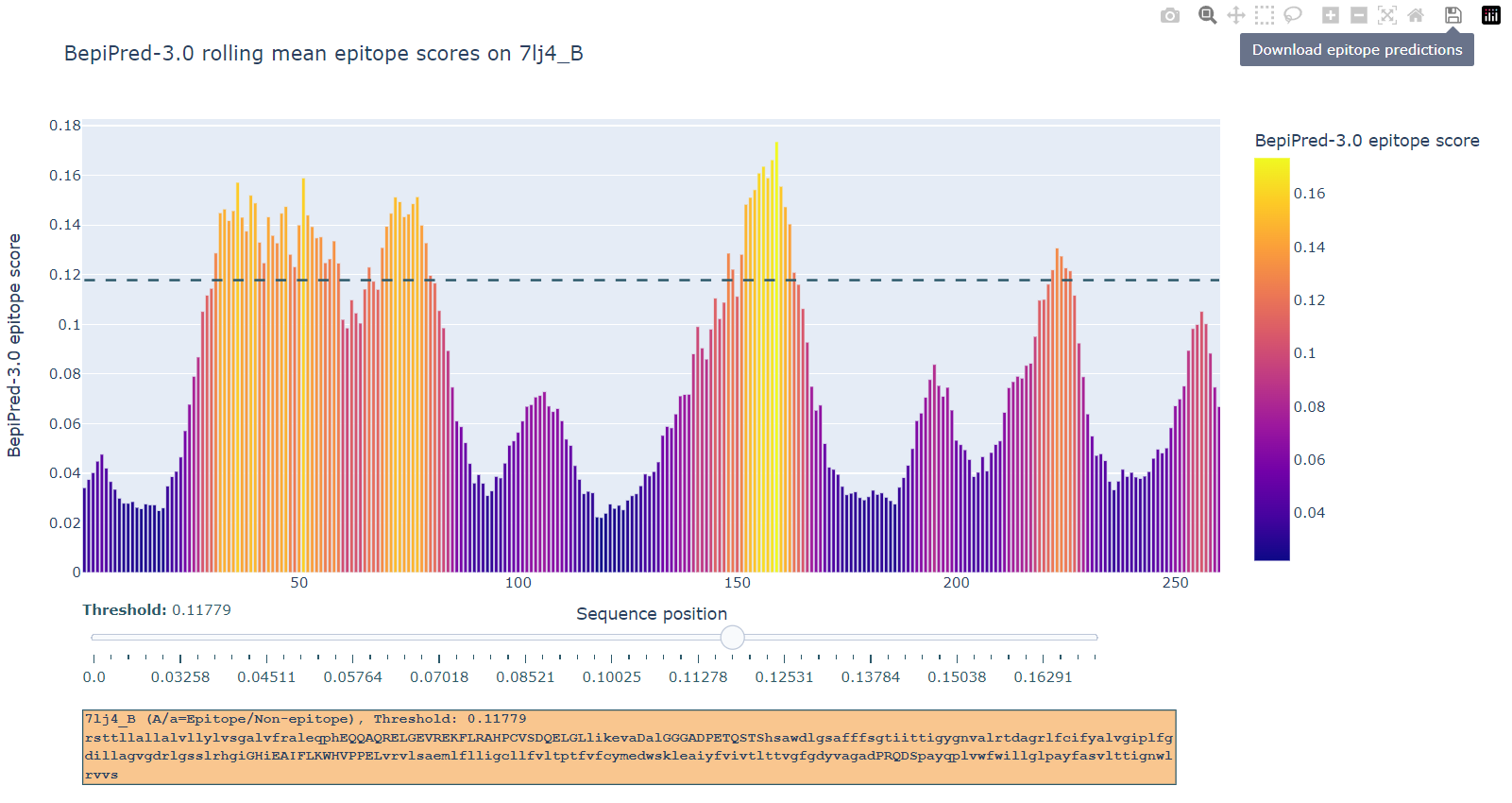
The BepiPred-3.0 server predicts both linear and discontinous B-cell epitopes from protein sequence, using neural networks trained on state of the art protein language embeddings of epitope and non-epitope amino acids determined from crystal structures. A sequential smoothing (rolling mean) can be optionally used.
The BepiPred-3.0 server requires protein sequence(s) in fasta format, and can not handle nucleic acid sequences.
Here, one can download the data used for training, testing and evaluating this method.
File Format: Fasta
Header: <Positive/Negative>ID_<IEDB_Epitope_ID>
Two datasets constructed from linear epitopes extracted from IEDB. The first contains 4072 sequences.
In the reduced dataset, sequences with more than 20% sequence identity to the BP3C50ID training set were removed, leaving 3560 sequences.
Epitope: Uppercased
Non-Epitope: Lowercased
Downloads:
IEDB Linear Epitope Data
IEDB Linear Epitope Data Reduced
File Format: Fasta
Header: <PDBID>_<Chain_ID>
These datasets were constructed from crystal structures, deposited in the the PDB database before 29/09/2021. Note that because of the epitope annotation strategy used to construct datasets (BP3_training set, BP3C50ID training set, BP3C50ID external test set), sequences may differ slightly from what is in the PDB database.
Epitope: Uppercased
Non-Epitope: Lowercased
Downloads:
BP3_without epitope_collapse
BP3_training set
BP3C50ID training set
BP3C50ID external test set
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: