DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
Paste or upload protein sequence(s) as fasta format to predict potential B-cell epitopes. Prediction can take a few minutes per sequence.
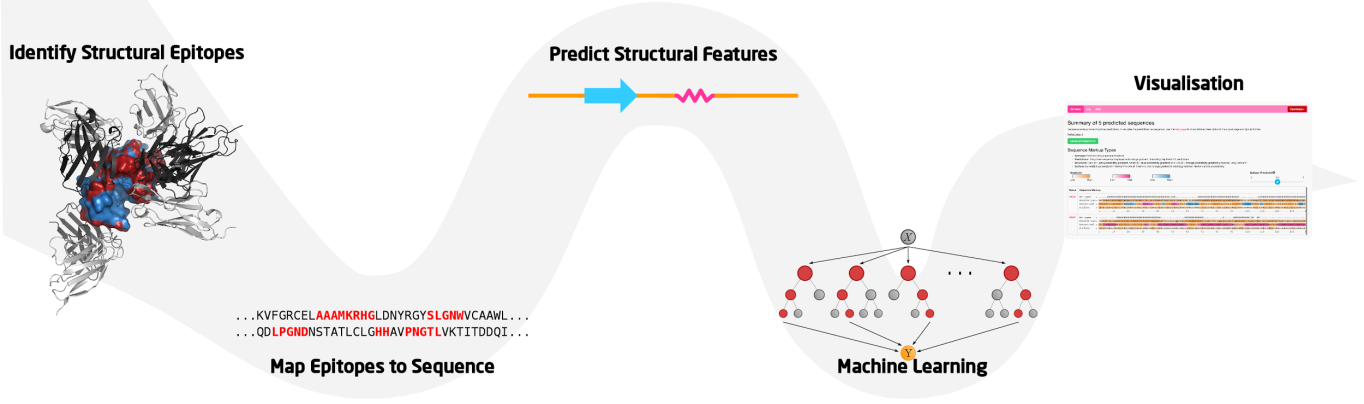
The BepiPred-2.0 server predicts B-cell epitopes from a protein sequence, using a Random Forest algorithm trained on epitopes and non-epitope amino acids determined from crystal structures. A sequential prediction smoothing is performed afterwards.
The BepiPred-2.0 server requires protein sequence(s) in fasta format, and can not handle nucleic acid sequences.
Paste protein sequence(s) in fasta format into field marked by arrow A or upload a fasta file marked by arrow B. Click the submit button, marked by arrow C, when protein sequences are entered.
After the server successfully finishes the job, a summary page shows up. If an error happens during modelling a log will appear specifying the error.
Use the navigation bar (arrow A) to flip through the various output pages. The default page is the summary, which shows a scrollable table (arrow D) illustrating the sequences with a sequence markup illustrating the B-cell epitope predictions. Hover over the sequence name to reveal the description of the sequence obtained from the fasta header.
Use the Epitope Threshold slider (arrow C) to interactively change the classified epitope residues, (E for epitopes and . for non-epitopes). For guidance of which Epitope Threshold is suitable for you, click the on the slider. Use the dropdown download button (arrow B) to download the output as JSON or CSV, or select "All Downloads" to get short description of the download files.
A more advanced output visualitation is available, if clicked the "Advanced Output is Off" button.
When advanced outformat is activated, predictions from NetsurfP is additionally available as sequence markup for each sequence and a description of the markup types are revealed (arrow A).
Two new gradients are added, illustrating to easily distinguish between secondary structure types. Coils, relative surface accessibility and BepiPred-2.0 epitope predictions all carry same gradient, as epitopes often are found in coils and needs to be exposed. This can be used to easily find correlations of high scoring areas between these three types.
Here, one can download the data used for training, testing and evaluating this method.
File Format: Fasta
Header: <Positive/Negative>ID_<IEDB_Epitope_ID>
The header explains whether the sequence has a negative or positive epitope sequence mapped onto it.
Epitope: Uppercased
Non-Epitope: Lowercased
Download: IEDB Linear Epitope Dataset
File Format: Fasta
Header: <PDBID>_<Heavy_Chain_ID><Light_Chain_ID> <Antigen_Chain_ID> <Partition>
The header explains which antibody chains and antigen chains in a given PDB have been used. The partition is which of the 5 randomly split partitions it belonged to and the partition EVAL is the completely left out PDBs.
Epitope: Uppercased
Non-Epitope: Lowercased
Download: PDB Combined Epitopes Dataset
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: