1. Sequencing data
Upload a FASTQ (or FASTA) file with the raw sequencing reads.
2. Barcode information
- Sample identification tag:
Upload a FASTA file with the sequences of the sample identification tags. The names of each sequence should correspond to the keys in the 3. Sample identification table.
- Forward primer A:
Paste the sequence of the forward primer from Oligo-A.
- N sequence:
Give an integer specifying the length of the first UMI (unique molecular identifier).
- Epitope tag A:
Upload a FASTA file with sequences of the 25mer oligonucleotide sequence from Oligo A.
The names of the FASTA entries must not include any spaces, and they must include "_Axx" at the end, where "xx" is an id number.
Example:
>Oligo_A1 CGAGGGCAATGGTTAACTGACACGT >Oligo_A2 CAGAAAGCAGTCTCGTCGGTTCGAA
Note that Barracoda also works for barcode designs that user other lengths than 25.
- Annealing region:
Paste the sequence of the annealing region.
- Epitope tag B:
Upload a FASTA file with sequences of the 25mer oligonucleotide sequence from Oligo B.
The sequences should be from reverse strand and orientation compared to the Oligo A sequences.
The names of the FASTA entries must not include any spaces, and they must include "_Byy" at the end, where "yy" is an id number.
Example:
>Oligo_B1 GCCTGTAGTCCCACGCGATCTAACA >Oligo_B2 CAACCATTGATTGGGGACAACTGGG
Note that Barracoda also works for barcode designs that user other lengths than 25.
- N sequence:
Give an integer specifying the length of the second UMI (unique molecular identifier).
- Forward primer B:
Paste the sequence of the reverse primer from Oligo-B. The sequence should be from reverse strand and orientation compared to the Primer A sequence.
3. Sample identification
Upload a tab-delimited file (with no headers), where each row is of the format:<key> <tab> <sample name> <tab> <experiment>Example:
A-Key_2OS_F1_01 sampleA 1 A-Key_2OS_F1_02 sampleB 1 A-Key_2OS_F1_03 sampleX 2 A-Key_2OS_F1_04 sampleY 2 A-Key_2OS_F1_05 input 1 A-Key_2OS_F1_06 input 1 A-Key_2OS_F1_07 input 2 A-Key_2OS_F1_08 input 2
-
Importantly, key should match the sequence names in the FASTA file uploaded in the field sample identification tag.
- Equally important, the sample name field should be "input" (all non-capital letters) for those samples that are used for controls.
- The sample name field will be used to annotate results and figures.
-
The experiment field is used to denote that the data should be treated as multiple experiments. The samples will be analysed in the groups defined by experiment. In the above example there are two experiments: sample A and B will be analysed together with the two first input samples. Sample X and Y will be analysed together with the two last input samples.
If all samples are to be analysed as one experiment, this field should simply contain the same value in all the rows (eg. "1").
4. Barcode annotations
NEW: file can also be a Microsoft Excel workbook, where each sheet contains an annotation table as described below. The name of each sheet must match the experiment names given in the sample identification table above.Upload a tab-delimited file with user-specified annotations for the DNA barcodes. The file should have headers, but there are no specific requirements for the headers.
- The first column should contain the AxBy name of each barcode made by combining Oligos A with Oligos B, e.g. "A1B2".
- There can be any number of additional columns with information about these barcodes, such as the peptide they are linked to, and this data will be appended to the output where possible (mainly in excel sheets.)
- Make sure there are no special symbols in the annotation file (such as greek letters, url links etc). You can easily remove such symbols with online copy-paste tools, such as this one.
Barcode HLA allele Peptide Sequence A7B1 HLA-A0201 707-AP RVAALARDAP A7B2 HLA-A0201 ATIC (AICRT) RLDFNLIRV A7B3 HLA-A0201 ATIC (AICRT) MVYDLYKTL
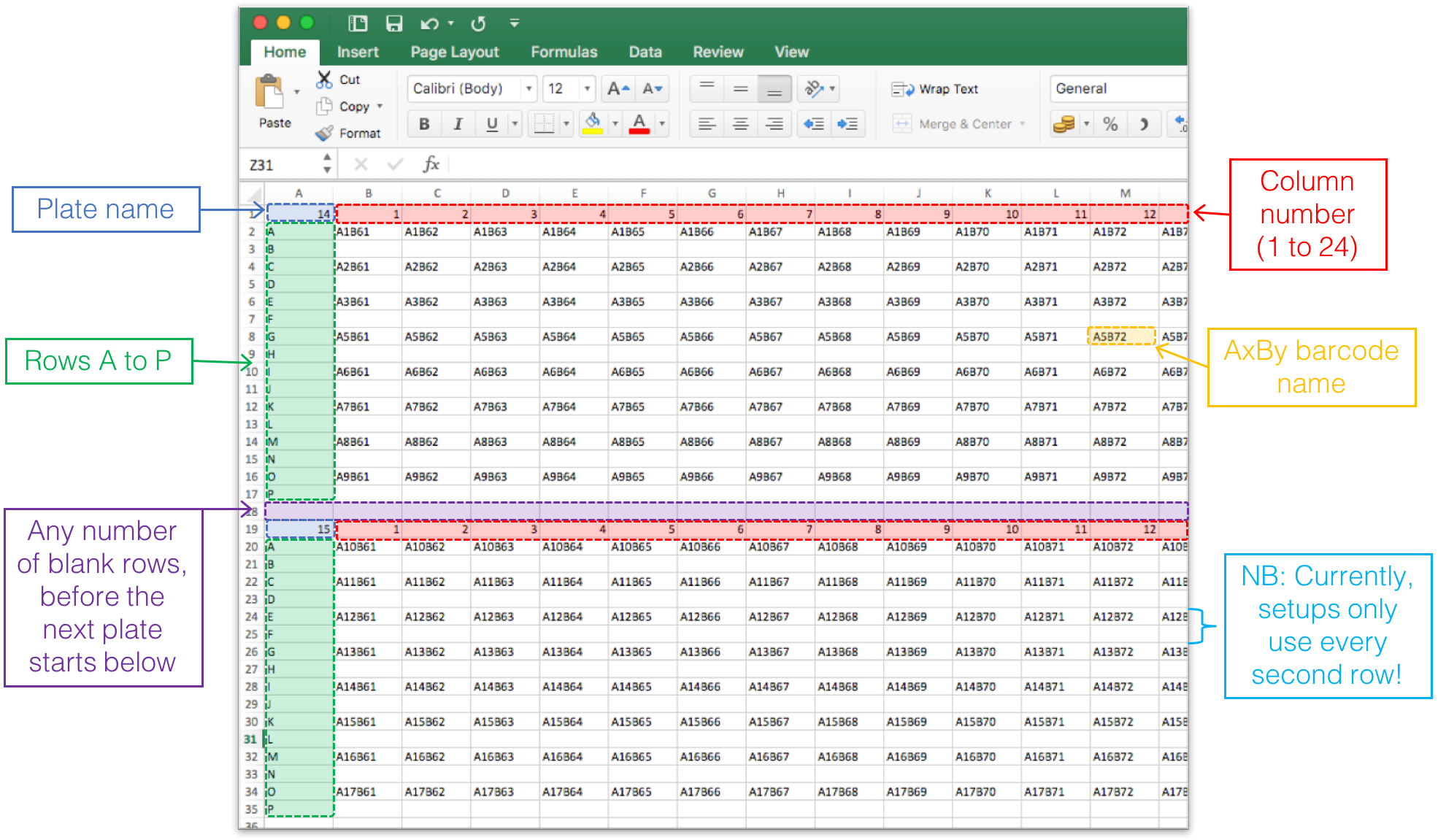
5. Barcode plate setups (optional)
Optionally, upload an excel file that describes how the DNA barcodes were layed out on 384-well plates in the experimental setup. This will be used to make figures of your results, where the values (read counts, log fold change, p-values) are layed out on a heatmap-type plot to mimic the 384-well plate. This will hopefully aid in detecting experimental errors and biases, such as spill-over between wells, or mix-up of barcodes on the plates.Example of excel sheet:
5. Submit the job
Click on the "Submit" button. The status of your job (either 'queued' or 'running') will be displayed and constantly updated until it terminates and the server output appears in the browser window.At any time during the wait you may enter your e-mail address and simply leave the window. Your job will continue; you will be notified by e-mail when it has terminated. The e-mail message will contain the URL under which the results are stored; they will remain on the server for 24 hours for you to collect them.