Instructions
AbEpiTope-1.0 is a tool that features two scores. The first is AbEpiScore-1.0, which is desgined to evaluate the accuracy of modelled antibody-antigen interfaces. The second is, AbEpiTarget-1.0, a score designed to distinguish AbAg complexes modelled with the correct antibody, from those modelled with incorrect or "swapped" antibodies.
Input
1. Users can upload a single structure file (pdb/cif) or a zip file containing pdb/cif files.
Each structure file must include a light and heavy chain or a single-chain variable fragment (scFv), along with one or more antigen chains. Due to computational resource limits on the web server, we restrict uploads to a maximum of 30 files per submission.
For larger batches, we recommend using the local installation (see Versions or Download)
Note: Scores will not be produced for antibody-antigen structures where this is not detected.
2. Users can set a custom Angstrom (Å) distance for defining antibody-antigen interfaces.
The default is 4 Å. The antibody-antigen interface is made up of epitope and paratope residues. Epitope residues are any residues with at least one heavy atom (main-chain or side-chain) at a distance of 4 Å or less to any light or heavy chain. The corresponding interacting residues on the light or heavy chain are the paratope residues.
Note: Scores will not be produced for antibody-antigen structures if no epitope and paratope residues are detected at the set Å distance.
Output
See Output format
Guidance on using AbEpiTope-1.0 scores for predicting modelled AbAg accuracy
and antibody screening
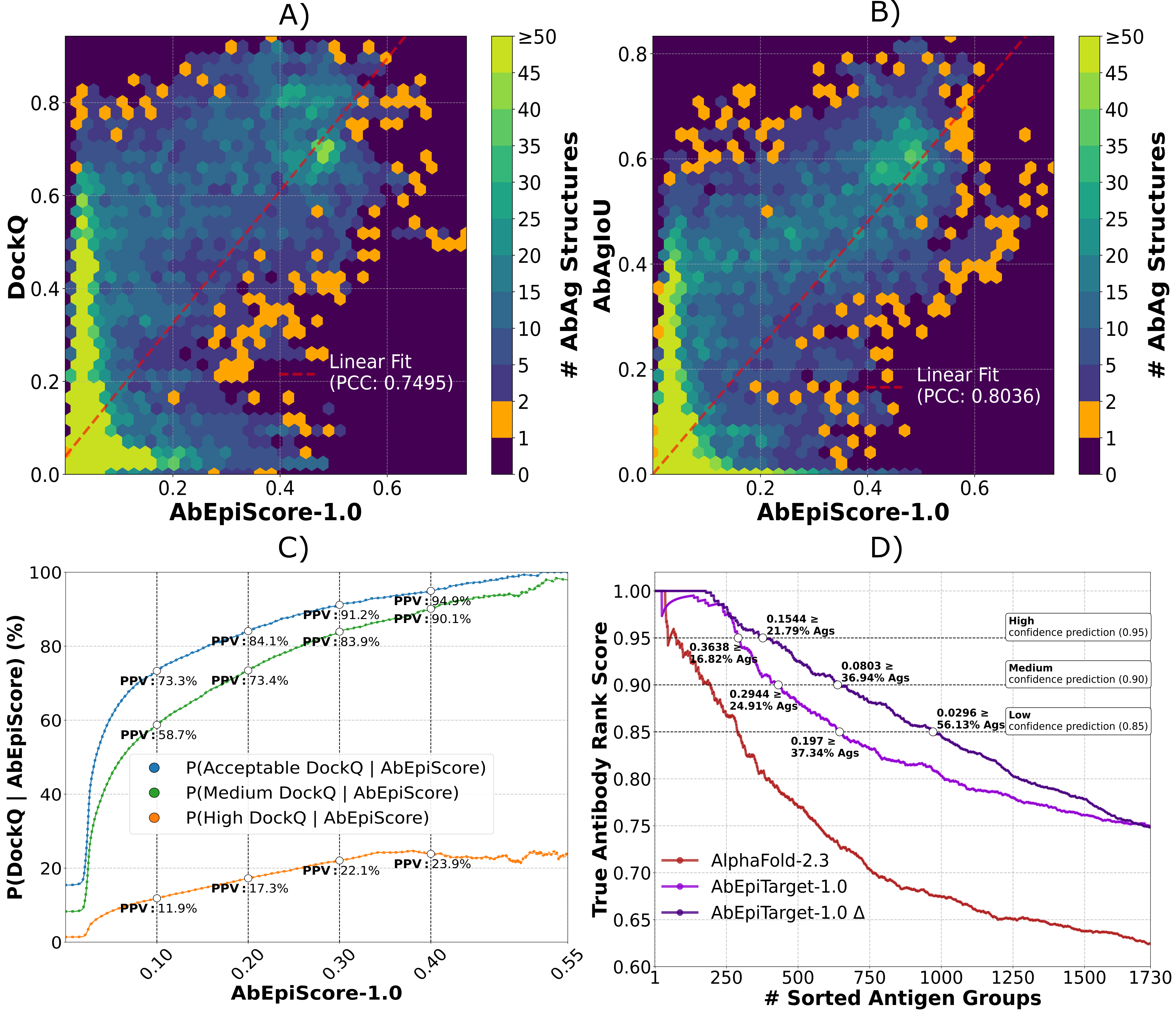
Figure: We illustrate how AbEpiScore-1.0 can be applied to predict the modelled AbAg accuracy and how AbEpiTarget-1.0 can be used for antibody screening A) AbEpiScore-1.0 scores for 51.900 predicted antibody-antigen structures plotted against corresponding DockQ values (y-axis) in hexagonal bins. Color scales capped at 50 structures, show the structure count per bin and orange indicates single structure bins. A red dashed line indicates a linear fit computed across all AbAg structures. B) Same as A), but plotting AbEpiScore-1.0 against AbAgIoU values. C) The PPV values for predicting whether 51.900 predicted antibody-antigen complexes have acceptable, medium or high DockQ (y-axis) as a function of X AbEpiScore-1.0 (x-axis) was computed in the range (0.0-0.55). PPV values for selected AbEpiScore are indicated with white dots. D) Evaluation of AbEpiTarget-1.0, AbEpiTarget-1.0 Δ, and AlphaFold 2.3 confidence scores for identifying correct AbAg pairs. For AbEpiTarget-1.0 and AlphaFold 2.3, the 1730 antigens groups were ranked by their maximum score (from either the true AbAg or one of the three swapped AbAgs). For AbEpiTarget-1.0 Δ, by the score gap between the top two pairs. The y-axis shows average True Rank Scores (0 = worst, 1 = perfect ranking of true AbAg) as more antigen groups are included (x-axis). White dots indicate score cutoffs corresponding to expected True Rank Scores of 0.85, 0.90, and 0.95.
- Predicting modelled AbAg Accuracy with AbEpiScore-1.0, A) and B): A modelled antibody-antigen structure with an AbEpiScore-1.0 of 0.3, has an expected interface accuracy 0.3 ≈ 0.400 DockQ or ≈0.373 AbAgIoU.
- Predicting modelled AbAg Accuracy with AbEpiScore-1.0, C): A modelled antibody-antigen structure with an AbEpiScore-1.0 of 0.3, has a 91.2%, 83.9%, and 22.1% probability of having acceptable (≥0.23), medium (≥0.49) and high (≥0.8) DockQ interface accuracy respectively.
- High confidence antibody screening with AbEpiTarget-1.0, D): For an antigen group (antibodies modelled to the same antigen), only predict the highest scoring antibody-antigen pair as the correct antibody, if its AbEpiTarget-1.0 score is 0.1544 more than the second best scoring antibody-antigen pair.
- Medium confidence antibody screening with AbEpiTarget-1.0, D): For an antigen group (antibodies modelled to the same antigen), only predict the highest scoring antibody-antigen pair as the correct antibody, if its AbEpiTarget-1.0 score is 0.0803 more than the second best scoring antibody-antigen pair.
- Low confidence antibody screening with AbEpiTarget-1.0, D): For an antigen group (antibodies modelled to the same antigen), only predict the highest scoring antibody-antigen pair as the correct antibody, if its AbEpiTarget-1.0 score is 0.0296 more than the second best scoring antibody-antigen pair.
Abstract
AbEpiTope-1.0: Improved antibody target prediction by use of AlphaFold and inverse folding
Authors: Joakim Clifford, Eve Richardson, Bjoern Peters, Morten Nielsen
Publication:
Abstract
B-cell epitope prediction tools are crucial for the design of vaccines and disease diagnostics. However, predicting which antigens a specific antibody will bind to, and their exact binding sites (epitopes), remains challenging. Here, we present AbEpiTope-1.0, a computational tool for antibody-specific B-cell epitope prediction, utilising AlphaFold-2.3 for structural modelling and inverse folding for the machine learning models. AbEpiTope-1.0 outperforms AlphaFold’s confidence ranking in predicting the accuracy of modelled antibody-antigen interfaces. Most importantly, we show that the predicted accuracy is sensitive to antibody input, offering a reliable metric for selecting antibodies most likely to bind a given antigen. Furthermore, a variant of our model trained specifically for this task shows a significant performance improvement. The tool can evaluate hundreds of antibody-antigen structures in minutes, providing researchers with a valuable resource for antibody screening and B-cell epitope prediction. AbEpiTope-1.0 is freely available as a web server and standalone package at https://services.healthtech.dtu.dk/services/AbEpiTope-1.0.
Predicting Antibody-Antigen Interface Accuracy
This data is related to predicting the interface accuracy of modelled AbAg structures. We first tested AbAg interface scores for AlphaFold-2.3 and inverse folding GVP-Transformers, ESMIF1 and AntiFold, on 1,730 AbAgs without fine-tuning, creating 30 structures for each using AlphaFold-2.3 multimer, totalling 51,900 structures. AbAgIoU was used to measure the match between predicted epitope and paratope residues and the corresponding ground truth crystal structures. Later, we created finetuned models: OneHot-AbAgIoU, ESM2-AbAgIoU, AntiFold-AbAgIoU and AbEpiScore-1.0.
Downloads
AlphaFold-2.3 (ColabFold) AbAg Fasta Inputs:
A .zip file with containing all input fasta files for modelling 1730 antibody-antigen complexes with AlphaFold-2.3 colabfold version. We used 6 seeds, generating 30 structures per antibody-antigen complex and 51900 structures in total.
File Format: .zip
Download: abag_fastafiles.zip
AbAg Interface Scores:
A .csv file with AbAgIoU and DockQ scores for all 51900 structures, as well as corresponding AbAg interface model scores done in nested cross-validation. These models were: Random, Onehot-AbAgIoU, ESM2-AbAgIoU, AlphaFold-2.3, AntiFold, AntiFold-AbAgIoU, ESMIF1, AbEpiScore-1.0. Nested cross-validation was done b creating 5 data partitions of antibody-antigen complexes not sharing more 65% or 95% antigen or antibody sequence identity. Data partitions are indicated by PartitionNum
File Format: .csv
Header: StructureNames,AbAgIoU,AgIoU,DockQ, AbAg Interface Scores..., PartitionNum
Download: abag_interface_scores.csv
Antibody Target Prediction
This data is related to predicting the antigen target of a given antibody, distinguishing modelled true AbAg complexes from those modelled with incorrect or "swapped" antibodies. All modelled AbAg complexex were made with AlphaFold-2.3. We created 1,730 groups of antibody-antigen complexes, each containing one true antibody-anitgen complex and three swapped antibody-antigen complexes, all modelled with the same antigen. To avoid data leakage, antibodies for constructing swap antibody-antigen complexes were taken from other antibody-antigen complexes within the same data partition and only if the antibody was targeting a different antigen.
AlphaFold-2.3 (ColabFold) Swap AbAg Fasta Inputs:
A .zip file with containing all input fasta files for modelling 1730x3 = 5190 swapped antibody-antigen complexes with AlphaFold-2.3 colabfold version. We used 6 seeds, generating 30 structures per antibody-antigen complex and 51900 structures in total.
File Format: .zip
Download: swap_abag_fastafiles.zip
Antibody Target Prediction Scores:
A .csv file with AgIoU scores, measuring the match between predicted epitope residues and the ground truth crystal structure epitopes, for the 51900 antibody-antigen complexes and the 155700 swapped antibody-antigen structures. We also supply the corresponding model scores all evaluated in nested cross-validation. These include AbAg interface score models as well as models made specifically for antibody target predcition. These models were: Random, AlphaFold-2.3, Onehot-AbAgIoU, AbEpiTarget-1.0 (OneHot), AbEpiTarget-1.0 (OneHot+AlphaFold-2.3), AbEpiTarget-1.0 (OneHot+AbEpiScore-1.0), ESM2-AbAgIoU, AbEpiTarget-1.0 (ESM2+AlphaFold-2.3), AbEpiTarget-1.0 (ESM2+AbEpiScore-1.0) ESM2-AbAgIoU, AntiFold, AntiFold-AbAgIoU, ESMIF1, AbEpiScore-1.0, AbEpiTarget-1.0 AbEpiTarget-1.0 (+AlphaFold-2.3) and AbEpiTarget-1.0 (+AbEpiScore-1.0). Data partitions are indicated by PartitionNum
File Format: .csv
Header: StructureNames,AgIoU,AbAg Target Scores...,PartitionNum
Download: abag_abtarget_scores.csv
Antibody Target Prediction - 17x17 swapped antibody-antigen complex dataset
This data is also related to predicting the antigen target of a given antibody. We assesed the model performances of antibody target prediction for scenarios with more than three swapped AbAgs. Here, we created 17 groups, each consisting of one true AbAg - featuring the correct antibody and antigen- as well as 16 swapped AbAgs, where the true antigen was paired with incorrect antibodies.
/net/urban/var/www/services/suppl/immunology/AbEpiTope-1.0/17x17_benchmark/abag_abtarget_scores.csv
AlphaFold-2.3 (ColabFold) True AbAg Fasta Inputs:
A .zip file with containing all input fasta files for modelling 17 antibody-antigen complexes with AlphaFold-2.3 colabfold version. We used 6 seeds, generating 30 structures per antibody-antigen complex and 510 structures in total.
File Format: .zip
Download: true_abag_fastafiles.zip
AlphaFold-2.3 (ColabFold) Swap AbAg Fasta Inputs:
A .zip file with containing all input fasta files for modelling 17 antibody-antigen complexes and 17x16= 272 swapped antibody-antigen complexes with AlphaFold-2.3 colabfold version. We used 6 seeds, generating 30 structures per antibody-antigen complex and 8160 structures in total.
File Format: .zip
Download: swap_abag_fastafiles.zip
Antibody Target Prediction Scores:
A .csv file with AgIoU scores, measuring the match between predicted epitope residues and the ground truth crystal structure epitopes, for the 510 antibody-antigen complexes and the 8160 swapped antibody-antigen structures. We also supply the corresponding AlphaFold-2.3 and AbEpiTarget-1.0 scores done in nested cross-validation.
File Format: .csv
Header: StructureNames,AgIoU,AlphaFold-2.3,AbEpiTarget-1.0
Download: abag_abtarget_scores.csv
GitHub
Please visit our GitHub repository for a local installment of current version
The code and data can be used freely by academic groups for non-commercial purposes.
If you plan to use these tools for any for-profit application, you are required to obtain a separate license (contact Morten Nielsen, morni@dtu.dk)
This service offers no downloadable software
See a list of available software
GitHub
Please visit our GitHub repository for a local installment of current version
The code and data can be used freely by academic groups for non-commercial purposes.
If you plan to use these tools for any for-profit application, you are required to obtain a separate license (contact Morten Nielsen, morni@dtu.dk)
