DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
DTU Health Tech
Department of Health Technology
This link is for the general contact of the DTU Health Tech institute.
If you need help with the bioinformatics programs, see the "Getting Help" section below the program.
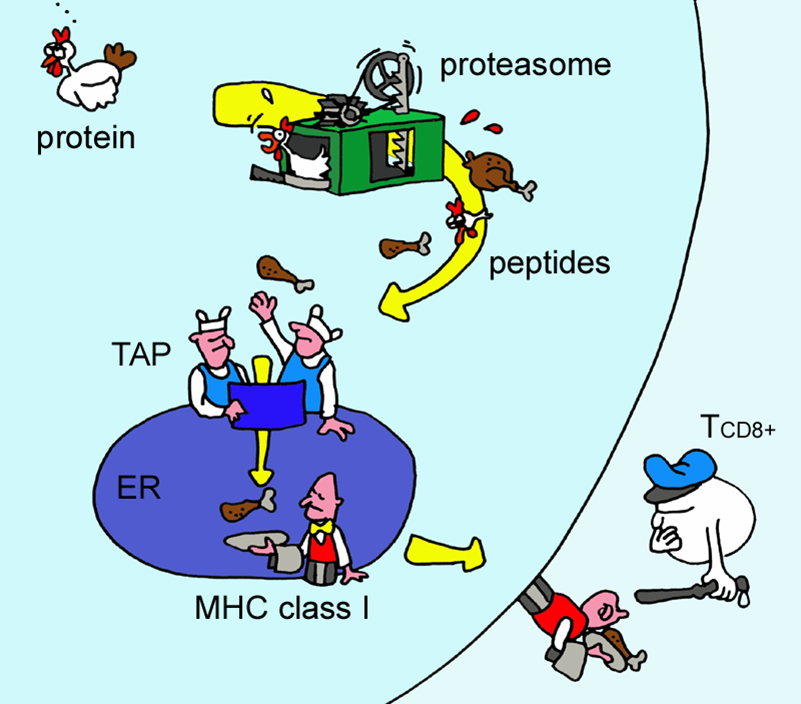 |
The NetCTL 1.2 server predicts CTL epitopes in protein sequences. The current version 1.2 is an update to the version 1.0. Version 1.2 expands the MHC class I binding predicition to 12 MHC supertypes including the supertypes A26 and B39. The accuracy of the MHC class I peptide binding affinity is significantly improved compared to the earlier version. Also the prediction of proteasonal cleavage has been improved and is now identical to the predictions obtained by the NetChop-3.0 server. The updated version has been trained on a set of 886 known MHC class I ligands.
NOTE. On Aug 16 2006 a minor update to the server has been implemented improving the prediction accuracy for MHC binding. The earlier version of the NetCTL 1.2 server (1.2 beta) is available via the versions history for the server.
The method integrates prediction of peptide MHC class I binding, proteasomal C terminal cleavage and TAP transport efficiency. The server allows for predictions of CTL epitopes restricted to 12 MHC class I supertype. MHC class I binding and proteasomal cleavage is performed using artificial neural networks. TAP transport efficiency is predictied using weight matrix.
The MHC peptide binding is predicted using neural networks trained as described for the NetMHC server. The proteasome cleavage event is predicted using the version of the NetChop neural networks trained on C terminals of known CTL epitopes as describe for the NetChop-3.0 server. The TAP transport efficiency is predicted using the weight matrix based method describe by Peters et al.
The server includes predictions of MHC/peptide binding for 12 MHC class I supertypes. The output from the neural network predicting MHC/peptide binding is a log transformed value related to the IC50 values in nM units. For details on the transformation please see the Output format
The scores from the three individual prediction methods are integrated as a weighted sum with a relative
weight on peptide/MHC binding of 1. Different thresholds for the integrated score can be translated into
sensitivity/specificity values. In a large benchmark calculate containing more than 800 known MHC class I ligands
the following relations were found
| Score | Sensitivity | Specificity |
| > 1.25 | 0.54 | 0.993 |
| > 1.00 | 0.70 | 0.985 |
| > 0.90 | 0.74 | 0.980 |
| > 0.75 | 0.80 | 0.970 |
| > 0.50 | 0.89 | 0.940 |
The project is collaboration between CBS and IMMI at Copenhagen university
For publication of results, please cite:
NetCTL-1.2:
Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction.
Larsen MV, Lundegaard C, Lamberth K, Buus S, Lund O, Nielsen M.
BMC Bioinformatics. Oct 31;8:424. 2007
View the abstract
NetCTL-1.0:
An integrative approach to CTL epitope prediction.
A combined algorithm integrating MHC-I binding, TAP transport efficiency, and proteasomal cleavage predictions.
Larsen M.V., Lundegaard C., Kasper Lamberth, Buus S,. Brunak S., Lund O., and Nielsen M.
European Journal of Immunology. 35(8): 2295-303. 2005
View the abstract
Reliable prediction of T-cell epitopes using neural networks with novel
sequence representations.
Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S,
Brunak S, Lund O.
Protein Sci., 12:1007-17, 2003.
View the abstract
Sensitive quantitative predictions of peptide-MHC binding by a 'Query by Committee' artificial neural network approach.
Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S.
Tissue Antigens., 62(5):378-84, 2003.
View the abstract
The role of the proteasome in generating cytotoxic T cell epitopes:
Insights obtained from improved predictions of proteasomal cleavage.
M. Nielsen, C. Lundegaard, S. Brunak, O. Lund, and C. Kesmir.
Immunogenetics., 57(1-2):33-41, 2005.
View the abstract
Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors
Peters, B., Bulik, S., Tampe, R., Endert, P. M. V. and Holzhutter, H. G.
J. Immunol. 171: 1741-1749, 2003.
View the abstract
The sequence must be written using the one letter
amino acid code:
`acdefghiklmnpqrstvwy' or
`ACDEFGHIKLMNPQRSTVWY'.
Other letters will be converted to `X' and treated as unknown
amino acids.
Other characters, such as whitespace and
numbers, will simply be ignored.
NetCTL predictions using MHC supertype A24. Threshold 0.500000 ....... 17 ID 143B_BOVIN pep AERYDDMAA aff 0.0349 aff_rescale 0.0478 cle 0.0344 tap -0.5270 COMB 0.0249 18 ID 143B_BOVIN pep ERYDDMAAA aff 0.0348 aff_rescale 0.0476 cle 0.4333 tap -0.1920 COMB 0.0813 19 ID 143B_BOVIN pep RYDDMAAAM aff 0.3017 aff_rescale 0.4128 cle 0.9943 tap 0.4700 COMB 0.5357 <-E 20 ID 143B_BOVIN pep YDDMAAAMK aff 0.0352 aff_rescale 0.0482 cle 0.9622 tap 0.0930 COMB 0.1491 21 ID 143B_BOVIN pep DDMAAAMKA aff 0.0343 aff_rescale 0.0469 cle 0.6469 tap -1.1460 COMB 0.0543 ...... ---------------------------- Number of MHC ligands 8 identified. Number of amino acids 237. Protein name 143B_BOVIN ----------------------------
SYFPEITHI dataset
HIV dataset
HIV_EpiJen dataset
List of known CTL epitopes from SYFPEITHI and the Los Alamos HIV databases
Difference between the A1, A2, and A3 epitope-protein pairs for the HIV and HIV_EpiJen datasets
Supplementary figure: Comparing specificities on the HIV EpiJen dataset
BACKGROUND: Reliable predictions of Cytotoxic T lymphocyte (CTL) epitopes are essential for rational vaccine design. Most importantly, they can minimize the experimental effort needed to identify epitopes. NetCTL is a web-based tool designed for predicting human CTL epitopes in any given protein. It does so by integrating predictions of proteasomal cleavage, TAP transport efficiency, and MHC class I affinity. At least four other methods have been developed recently that likewise attempt to predict CTL epitopes: EpiJen, MAPPP, MHC-pathway, and WAPP. In order to compare the performance of prediction methods, objective benchmarks and standardized performance measures are needed. Here, we develop such large-scale benchmark and corresponding performance measures and report the performance of an updated version 1.2 of NetCTL in comparison with the four other methods.
RESULTS: We define a number of performance measures that can handle the different types of output data from the five methods. We use two evaluation datasets consisting of known HIV CTL epitopes and their source proteins. The source proteins are split into all possible 9 mers and except for annotated epitopes; all other 9 mers are considered non-epitopes. In the RANK measure, we compare two methods at a time and count how often each of the methods rank the epitope highest. In another measure, we find the specificity of the methods at three predefined sensitivity values. Lastly, for each method, we calculate the percentage of known epitopes that rank within the 5% peptides with the highest predicted score.
CONCLUSION: NetCTL-1.2 is demonstrated to
have a higher predictive performance than EpiJen, MAPPP, MHC-pathway,
and WAPP on all performance measures. The higher performance of
NetCTL-1.2 as compared to EpiJen and MHC-pathway is, however, not
statistically significant on all measures. In the large-scale benchmark
calculation consisting of 216 known HIV epitopes covering all 12
recognized HLA supertypes, the NetCTL-1.2 method was shown to have a
sensitivity among the 5% top-scoring peptides above 0.72. On this
dataset, the best of the other methods achieved a sensitivity of 0.64.
The NetCTL-1.2 method is available at
http://services.healthtech.dtu.dk/service.php?NetCTL
All used datasets are available
at http://services.healthtech.dtu.dk/suppl/immunology/CTL-1.2.php
If you need help regarding technical issues (e.g. errors or missing results) contact Technical Support. Please include the name of the service and version (e.g. NetPhos-4.0) and the options you have selected. If the error occurs after the job has started running, please include the JOB ID (the long code that you see while the job is running).
If you have scientific questions (e.g. how the method works or how to interpret results), contact Correspondence.
Correspondence:
Technical Support: